4E95
 
 | | Lambda-[Ru(TAP)2(dppz-(Me)2)]2+ bound to CCGGATCCGG | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), 5'-D(*CP*CP*GP*GP*AP*TP*CP*CP*GP*G)-3', BARIUM ION | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | The effects of disubstitution on the binding of ruthenium complexes to DNA
Thesis, University of Reading, 2014
|
|
8TE7
 
 | | Structure of TRNM-f.01 | | Descriptor: | TRNM-f.01 Fab Heavy Chain, TRNM-f.01 Fab Light Chain | | Authors: | Bender, M.F, Olia, A.S, Kwong, P.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Broad and Potent HIV-1 Neutralization in Fusion Peptide-primed SHIV-boosted Macaques
To Be Published
|
|
1FBO
 
 | | Crystal structure of the cellulase CEL48F from C. cellulolyticum in complex with cellobiitol | | Descriptor: | CALCIUM ION, ENDO-1,4-BETA-GLUCANASE F, alpha-D-glucopyranose-(1-4)-D-glucose | | Authors: | Parsiegla, G, Reverbel-Leroy, C, Tardif, C, Belaich, J.P, Driguez, H, Haser, R. | | Deposit date: | 2000-07-16 | | Release date: | 2000-08-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Cellulase Cel48F in Complex with Inhibitors and Substrates Give Insights Into its Processive Action
Biochemistry, 39, 2000
|
|
5TPA
 
 | | Structure of the human GluN1/GluN2A LBD in complex with compound 9 (GNE3500) | | Descriptor: | (1R,2R)-2-(2-{[5-chloro-3-(trifluoromethyl)-1H-pyrazol-1-yl]methyl}-7-methyl-4-oxo-4H-pyrido[1,2-a]pyrimidin-6-yl)cyclopropane-1-carbonitrile, GLUTAMIC ACID, GLYCINE, ... | | Authors: | Wallweber, H.J.A, Lupardus, P.J. | | Deposit date: | 2016-10-20 | | Release date: | 2016-11-30 | | Last modified: | 2017-02-01 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | GluN2A-Selective Pyridopyrimidinone Series of NMDAR Positive Allosteric Modulators with an Improved in Vivo Profile.
ACS Med Chem Lett, 8, 2017
|
|
1F90
 
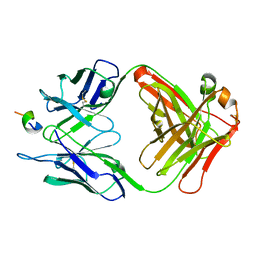 | | FAB FRAGMENT OF MONOCLONAL ANTIBODY (LNKB-2) AGAINST HUMAN INTERLEUKIN-2 IN COMPLEX WITH ANTIGENIC PEPTIDE | | Descriptor: | ANTIGENIC NONAPEPTIDE, FAB FRAGMENT OF MONOCLONAL ANTIBODY | | Authors: | Afonin, P.V, Fokin, A.V, Tsigannik, I.N, Mikhailova, I.Y, Onoprienko, L.V, Mikhaleva, I.I, Ivanov, V.T, Mareeva, T.Y, Nesmeyanov, V.A, Li, N, Duax, W.L, Pletnev, V.Z. | | Deposit date: | 2000-07-06 | | Release date: | 2001-07-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of an anti-interleukin-2 monoclonal antibody Fab complexed with an antigenic nonapeptide.
Protein Sci., 10, 2001
|
|
8PQJ
 
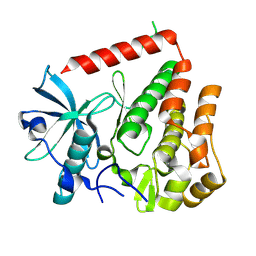 | | PDGFRA wild-type kinase domain | | Descriptor: | Platelet-derived growth factor receptor alpha | | Authors: | Teuber, A, Mueller, M.P, Rauh, D. | | Deposit date: | 2023-07-11 | | Release date: | 2023-12-27 | | Last modified: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Avapritinib-based SAR studies unveil a binding pocket in KIT and PDGFRA.
Nat Commun, 15, 2024
|
|
5MUF
 
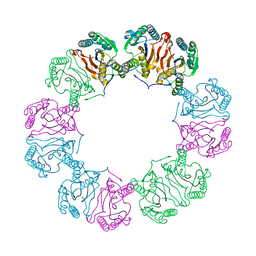 | | Crystal structure of human phosphoglycerate mutase family member 5 (PGAM5) in its enzymatically active dodecameric form induced by the presence of the N-terminal WDPNWD motif | | Descriptor: | PHOSPHATE ION, Serine/threonine-protein phosphatase PGAM5, mitochondrial | | Authors: | Chaikuad, A, Alfano, I, Picaud, S, Filippakopoulos, P, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-01-13 | | Release date: | 2017-07-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of PGAM5 Provide Insight into Active Site Plasticity and Multimeric Assembly.
Structure, 25, 2017
|
|
6LAN
 
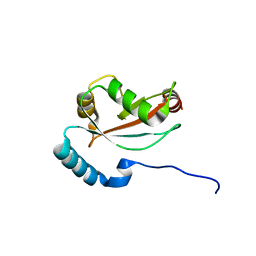 | | Structure of CCDC50 and LC3B complex | | Descriptor: | Coiled-coil domain-containing protein 50,Microtubule-associated proteins 1A/1B light chain 3B | | Authors: | Liu, L, Li, J, Hou, P. | | Deposit date: | 2019-11-12 | | Release date: | 2020-09-30 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | A novel selective autophagy receptor, CCDC50, delivers K63 polyubiquitination-activated RIG-I/MDA5 for degradation during viral infection.
Cell Res., 31, 2021
|
|
7VP2
 
 | | Structure of a transcription factor and DNA complex | | Descriptor: | DNA (5'-D(*AP*TP*GP*TP*GP*GP*TP*CP*CP*CP*CP*AP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*TP*GP*GP*GP*GP*AP*CP*CP*AP*CP*A)-3'), Transcription factor TCP10 | | Authors: | Zhang, Y, Xu, Y.P, Wang, B, Su, X.D. | | Deposit date: | 2021-10-15 | | Release date: | 2022-10-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural basis for DNA recognition by TCP transcription factors
To Be Published
|
|
1JE8
 
 | | Two-Component response regulator NarL/DNA Complex: DNA Bending Found in a High Affinity Site | | Descriptor: | 5'-D(*CP*GP*TP*AP*CP*CP*CP*AP*TP*TP*AP*AP*TP*GP*GP*GP*TP*AP*CP*G)-3', Nitrate/Nitrite Response Regulator Protein NARL, SULFATE ION | | Authors: | Maris, A.E, Sawaya, M.R, Kaczor-Grzeskowiak, M, Jarvis, M.R, Bearson, S.M.D, Kopka, M.L, Schroder, I, Gunsalus, R.P, Dickerson, R.E. | | Deposit date: | 2001-06-15 | | Release date: | 2002-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Dimerization allows DNA target site recognition by the NarL response regulator.
Nat.Struct.Biol., 9, 2002
|
|
4E8S
 
 | | Lambda-[Ru(TAP)2(dppz{Me2}2)]2+ bound to TCGGCGCCGA at high resolution | | Descriptor: | (11,12-dimethyldipyrido[3,2-a:2',3'-c]phenazine-kappa~2~N~4~,N~5~)[bis(pyrazino[2,3-f]quinoxaline-kappa~2~N~1~,N~10~)]ruthenium(2+), 5'-D(*TP*CP*GP*GP*CP*GP*CP*CP*GP*A)-3', BARIUM ION, ... | | Authors: | Hall, J.P, Cardin, C.J. | | Deposit date: | 2012-03-20 | | Release date: | 2013-03-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | The Structural Effect of Methyl Substitution on the Binding of Polypyridyl Ru dppz Complexes to DNA
Organometallics, 2015
|
|
1FBL
 
 | | STRUCTURE OF FULL-LENGTH PORCINE SYNOVIAL COLLAGENASE (MMP1) REVEALS A C-TERMINAL DOMAIN CONTAINING A CALCIUM-LINKED, FOUR-BLADED BETA-PROPELLER | | Descriptor: | CALCIUM ION, FIBROBLAST (INTERSTITIAL) COLLAGENASE (MMP-1), N-[3-(N'-HYDROXYCARBOXAMIDO)-2-(2-METHYLPROPYL)-PROPANOYL]-O-TYROSINE-N-METHYLAMIDE, ... | | Authors: | Li, J, Brick, P, Blow, D.M. | | Deposit date: | 1995-04-24 | | Release date: | 1996-01-29 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of full-length porcine synovial collagenase reveals a C-terminal domain containing a calcium-linked, four-bladed beta-propeller.
Structure, 3, 1995
|
|
3JQ5
 
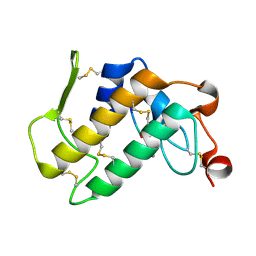 | | Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution | | Descriptor: | Amyloid Beta Peptide, CALCIUM ION, Phospholipase A2 isoform 3 | | Authors: | Mirza, Z, Vikram, G, Singh, N, Sinha, M, Bhushan, A, Sharma, S, Srinivasan, A, Kaur, P, Singh, T.P. | | Deposit date: | 2009-09-06 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Phospholipase A2 Prevents the Aggregation of Amyloid Beta Peptides: Crystal Structure of the Complex of Phospholipase A2 with Octapeptide Fragment of Amyloid Beta Peptide, Asp-Ala-Glu-Phe-Arg-His-Asp-Ser at 2 A Resolution
To be Published
|
|
2VAH
 
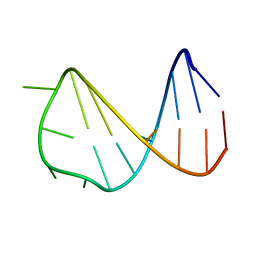 | | Solution structure of a B-DNA hairpin at low pressure. | | Descriptor: | 5'-D(*AP*GP*GP*AP*TP*CP*CP*TP*UP*TP *TP*GP*GP*AP*TP*CP*CP*T)-3' | | Authors: | Williamson, M.P, Wilton, D.J, Ghosh, M, Chary, K.V.A, Akasaka, K. | | Deposit date: | 2007-08-31 | | Release date: | 2007-09-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural Change in a B-DNA Helix with Hydrostatic Pressure
Nucleic Acids Res., 36, 2008
|
|
2UZ5
 
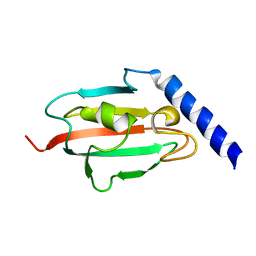 | | Solution structure of the fkbp-domain of Legionella pneumophila Mip | | Descriptor: | MACROPHAGE INFECTIVITY POTENTIATOR | | Authors: | Ceymann, A, Horstmann, M, Ehses, P, Schweimer, K, Steinert, M, Kamphausen, T, Fischer, G, Hacker, J, Rosch, P, Faber, C. | | Deposit date: | 2007-04-25 | | Release date: | 2007-06-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Domain Motions of the Mip Protein from Legionella Pneumophila
Biochemistry, 45, 2006
|
|
8PQK
 
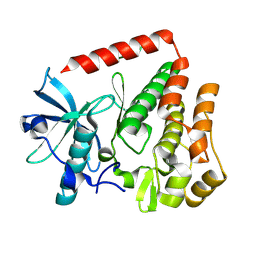 | |
4PZX
 
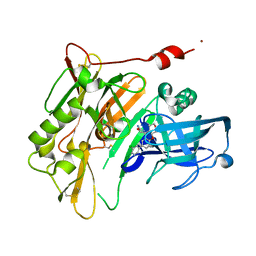 | | Synthesis, Characterization and PK/PD Studies of a Series of Spirocyclic Pyranochromene BACE1 Inhibitors | | Descriptor: | (4R,4a'R,10a'R)-7'-(5-chloropyridin-3-yl)-3',4',4a',10a'-tetrahydro-1'H-spiro[1,3-oxazole-4,5'-pyrano[3,4-b]chromen]-2-amine, Beta-secretase 1, NICKEL (II) ION | | Authors: | Vigers, G.P.A. | | Deposit date: | 2014-03-31 | | Release date: | 2014-05-14 | | Last modified: | 2014-10-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Synthesis, characterization, and PK/PD studies of a series of spirocyclic pyranochromene BACE1 inhibitors.
Bioorg.Med.Chem.Lett., 24, 2014
|
|
3JA6
 
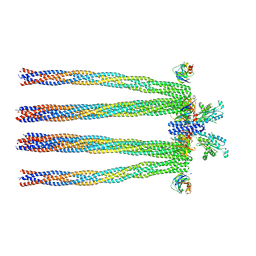 | | Cryo-electron Tomography and All-atom Molecular Dynamics Simulations Reveal a Novel Kinase Conformational Switch in Bacterial Chemotaxis Signaling | | Descriptor: | Chemotaxis protein CheA, Chemotaxis protein CheW, Methyl-accepting chemotaxis protein 2 | | Authors: | Cassidy, C.K, Himes, B.A, Alvarez, F.J, Ma, J, Zhao, G, Perilla, J.R, Schulten, K, Zhang, P. | | Deposit date: | 2015-04-21 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (12.7 Å) | | Cite: | CryoEM and computer simulations reveal a novel kinase conformational switch in bacterial chemotaxis signaling.
Elife, 4, 2015
|
|
1NDE
 
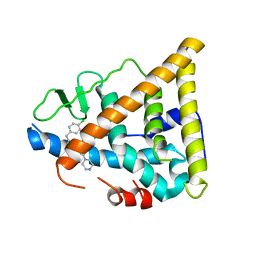 | | Estrogen Receptor beta with Selective Triazine Modulator | | Descriptor: | 4-(2-{[4-{[3-(4-CHLOROPHENYL)PROPYL]SULFANYL}-6-(1-PIPERAZINYL)-1,3,5-TRIAZIN-2-YL]AMINO}ETHYL)PHENOL, Estrogen receptor beta | | Authors: | Henke, B.R, Consler, T.G, Go, N, Hale, R.L, Hohman, D.R, Jones, S.A, Lu, A.T, Moore, L.B, Moore, J.T, Orband-Miller, L.A, Robinett, R.G, Shearin, J, Spearing, P.K, Stewart, E.L, Turnbull, P.S, Weaver, S.L, Williams, S.P, Wisely, G.B, Lambert, M.H. | | Deposit date: | 2002-12-09 | | Release date: | 2002-12-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A New Series of Estrogen Receptor Modulators That Display Selectivity for Estrogen Receptor beta
J.Med.Chem., 45, 2002
|
|
5FRA
 
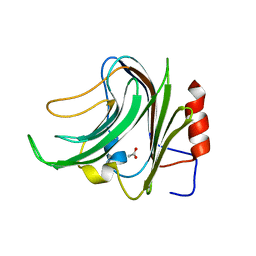 | | CBM40_CPF0721-6'SL | | Descriptor: | ACETATE ION, N-acetyl-alpha-neuraminic acid, N-acetyl-alpha-neuraminic acid-(2-6)-beta-D-galactopyranose, ... | | Authors: | Ribeiro, J.P, Pau, W, Pifferi, C, Renaudet, O, Varrot, A, Mahal, L.K, Imberty, A. | | Deposit date: | 2015-12-16 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of a High-Affinity Sialic Acid-Specific Cbm40 from Clostridium Perfringens and Engineering of a Divalent Form.
Biochem.J., 473, 2016
|
|
5MOA
 
 | | ABA RECEPTOR FROM TOMATO, SlPYL1 | | Descriptor: | SlPYL1 | | Authors: | Moreno-Alvero, M, Yunta, C, Gonzalez-Guzman, M, Arbona, V, Granell, A, Martinez-Ripoll, M, Infantes, L, Rodriguez, P.L, Albert, A. | | Deposit date: | 2016-12-14 | | Release date: | 2017-08-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure of Ligand-Bound Intermediates of Crop ABA Receptors Highlights PP2C as Necessary ABA Co-receptor.
Mol Plant, 10, 2017
|
|
6IY5
 
 | | NMR solution structures of 5'-ATTCTATTCT-3 | | Descriptor: | DNA (5'-D(*AP*TP*TP*CP*TP*AP*TP*TP*CP*T)-3'), SODIUM ION | | Authors: | Lam, S.L, Guo, P. | | Deposit date: | 2018-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Minidumbbell structures formed by ATTCT pentanucleotide repeats in spinocerebellar ataxia type 10.
Nucleic Acids Res., 48, 2020
|
|
1WTT
 
 | |
4XZ1
 
 | | ZAP-70-tSH2:Compound-B adduct | | Descriptor: | 2-[(7-chloro-4-nitro-2,1,3-benzoxadiazol-5-yl)amino]ethanol, Tyrosine-protein kinase ZAP-70, doubly phosphorylated ITAM peptide | | Authors: | Barros, T, Kuriyan, J, Winger, J.A, Visperas, P.R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-07-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Modification by covalent reaction or oxidation of cysteine residues in the tandem-SH2 domains of ZAP-70 and Syk can block phosphopeptide binding.
Biochem. J., 465, 2015
|
|
5F7L
 
 | | Blood group antigen binding adhesin BabA of Helicobacter pylori strain 17875 in complex with Nanobody Nb-ER14 | | Descriptor: | Adhesin binding fucosylated histo-blood group antigen, Nanobody Nb-ER14, SULFATE ION | | Authors: | Moonens, K, Gideonsson, P, Subedi, S, Romao, E, Oscarson, S, Muyldermans, S, Boren, T, Remaut, H. | | Deposit date: | 2015-12-08 | | Release date: | 2016-01-20 | | Last modified: | 2016-01-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Insights into Polymorphic ABO Glycan Binding by Helicobacter pylori.
Cell Host Microbe, 19, 2016
|
|
