5L4J
 
 | | Crystal Structure of Human Transthyretin in Complex with 4,4'-Dihydroxydiphenyl sulfone (Bisphenol S, BPS) | | Descriptor: | 4-(4-hydroxyphenyl)sulfonylphenol, SODIUM ION, Transthyretin | | Authors: | Grundstrom, C, Hall, M, Zhang, J, Olofsson, A, Andersson, P, Sauer-Eriksson, A.E. | | Deposit date: | 2016-05-25 | | Release date: | 2016-10-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Based Virtual Screening Protocol for in Silico Identification of Potential Thyroid Disrupting Chemicals Targeting Transthyretin.
Environ. Sci. Technol., 50, 2016
|
|
2ZXB
 
 | | alpha-L-fucosidase complexed with inhibitor, ph-6FNJ | | Descriptor: | (2S,3R,4S,5R)-2-benzylpiperidine-3,4,5-triol, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
3RYX
 
 | | Fluoroalkyl and Alkyl Chains Have Similar Hydrophobicities in Binding to the Hydrophobic Wall of Carbonic Anhydrase | | Descriptor: | Carbonic anhydrase 2, N-(2,2,3,3,3-pentafluoropropyl)-4-sulfamoylbenzamide, ZINC ION | | Authors: | Snyder, P.W, Bai, S, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-11 | | Release date: | 2011-08-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Fluoroalkyl and alkyl chains have similar hydrophobicities in binding to the "hydrophobic wall" of carbonic anhydrase.
J.Am.Chem.Soc., 133, 2011
|
|
7FIY
 
 | | Cryo-EM structure of the tirzepatide-bound human GIPR-Gs complex | | Descriptor: | CHOLESTEROL, Gastric inhibitory polypeptide receptor,Gastric inhibitory polypeptide receptor,human glucose-dependent insulinotropic polypeptide receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Zhao, F.H, Zhou, Q.T, Cong, Z.T, Hang, K.N, Zou, X.Y, Zhang, C, Chen, Y, Dai, A.T, Liang, A.Y, Ming, Q.Q, Wang, M, Chen, L.N, Xu, P.Y, Chang, R.L, Feng, W.B, Xia, T, Zhang, Y, Wu, B.L, Yang, D.H, Zhao, L.H, Xu, H.E, Wang, M.W. | | Deposit date: | 2021-08-01 | | Release date: | 2022-03-02 | | Last modified: | 2022-03-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural insights into multiplexed pharmacological actions of tirzepatide and peptide 20 at the GIP, GLP-1 or glucagon receptors.
Nat Commun, 13, 2022
|
|
5IED
 
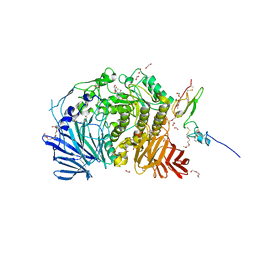 | | Murine endoplasmic reticulum alpha-glucosidase II with castanospermine | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-02-25 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3VZZ
 
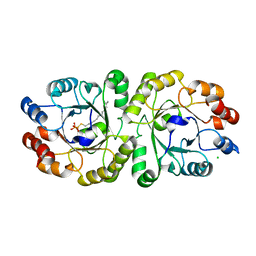 | | Crystal structure of PcrB complexed with FsPP from bacillus subtilis subap. subtilis str. 168 | | Descriptor: | CHLORIDE ION, Heptaprenylglyceryl phosphate synthase, MAGNESIUM ION, ... | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3A0M
 
 | | Structure of (PPG)4-OVG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Mizuno, K, Bachinger, H.P. | | Deposit date: | 2009-03-21 | | Release date: | 2010-03-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Stabilization of triple-helical structures of collagen peptides containing a Hyp-Thr-Gly, Hyp-Val-Gly, or Hyp-Ser-Gly sequence.
Biopolymers, 95, 2011
|
|
7SR0
 
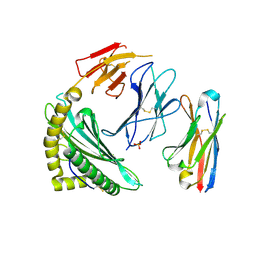 | | Single chain trimer HLA-A*02:01 (H98L, Y108C) with HPV.16 E7 peptide YMLDLQPET | | Descriptor: | PHOSPHATE ION, Protein E7 peptide,Beta-2-microglobulin,MHC class I antigen chimera, VHH | | Authors: | Finton, K.A.K, Rupert, P.B. | | Deposit date: | 2021-11-07 | | Release date: | 2022-11-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Effects of HLA single chain trimer design on peptide presentation and stability.
Front Immunol, 14, 2023
|
|
5IOP
 
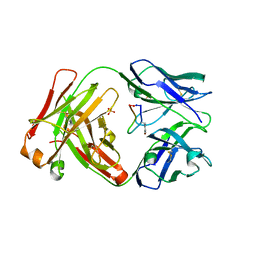 | |
5LGZ
 
 | | Structure of Photoreduced Pentaerythritol Tetranitrate Reductase | | Descriptor: | 1-DEOXY-1-(7,8-DIMETHYL-2,4-DIOXO-3,4-DIHYDRO-2H-BENZO[G]PTERIDIN-1-ID-10(5H)-YL)-5-O-PHOSPHONATO-D-RIBITOL, ISOPROPYL ALCOHOL, Pentaerythritol tetranitrate reductase | | Authors: | Kwon, H, Smith, O.M, Moody, P.C.E. | | Deposit date: | 2016-07-08 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Combining X-ray and neutron crystallography with spectroscopy.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
2MZG
 
 | | Purotoxin-2 NMR structure in DPC micelles | | Descriptor: | Purotoxin-2 | | Authors: | Nadezhdin, K, Oparin, P, Vassilevski, A, Grishin, E, Arseniev, A. | | Deposit date: | 2015-02-12 | | Release date: | 2016-04-13 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structure of purotoxin-2 from wolf spider: modular design and membrane-assisted mode of action in arachnid toxins.
Biochem. J., 473, 2016
|
|
1PRT
 
 | | THE CRYSTAL STRUCTURE OF PERTUSSIS TOXIN | | Descriptor: | PERTUSSIS TOXIN (SUBUNIT S1), PERTUSSIS TOXIN (SUBUNIT S2), PERTUSSIS TOXIN (SUBUNIT S3), ... | | Authors: | Stein, P.E, Read, R.J. | | Deposit date: | 1993-11-22 | | Release date: | 1995-01-26 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The crystal structure of pertussis toxin.
Structure, 2, 1994
|
|
1Q7A
 
 | | Crystal structure of the complex formed between russell's viper phospholipase A2 and an antiinflammatory agent oxyphenbutazone at 1.6A resolution | | Descriptor: | 4-BUTYL-1-(4-HYDROXYPHENYL)-2-PHENYLPYRAZOLIDINE-3,5-DIONE, METHANOL, Phospholipase A2 VRV-PL-VIIIa, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Singh, T.P. | | Deposit date: | 2003-08-17 | | Release date: | 2004-05-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Phospholipase A2 as a target protein for nonsteroidal anti-inflammatory drugs (NSAIDS): crystal structure of the complex formed between phospholipase A2 and oxyphenbutazone at 1.6 A resolution.
Biochemistry, 43, 2004
|
|
5LK1
 
 | |
3S77
 
 | | The origin of the hydrophobic effect in the molecular recognition of arylsulfonamides by carbonic anhydrase | | Descriptor: | 1,3-thiazole-2-sulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Snyder, P.W, Heroux, A, Whitesides, G.W. | | Deposit date: | 2011-05-26 | | Release date: | 2011-10-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Mechanism of the hydrophobic effect in the biomolecular recognition of arylsulfonamides by carbonic anhydrase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ZXA
 
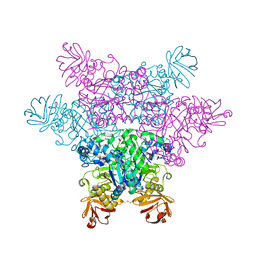 | | alpha-L-fucosidase complexed with inhibitor, FNJ-acetyl | | Descriptor: | Alpha-L-fucosidase, putative, N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}acetamide | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-22 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
5LJX
 
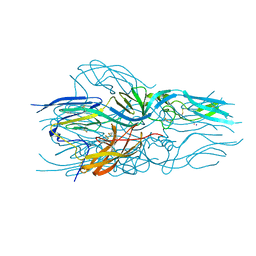 | |
5LKJ
 
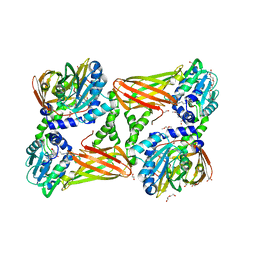 | | Crystal structure of mouse CARM1 in complex with ligand SA684 | | Descriptor: | (2~{S})-4-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl-(2-carbamimidamidoethyl)amino]-2-azanyl-butanoic acid, 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, ... | | Authors: | Cura, V, Marechal, N, Mailliot, J, Troffer-Charlier, N, Hassenboehler, P, Wurtz, J.M, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-07-22 | | Release date: | 2017-08-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.595 Å) | | Cite: | Crystal structure of mouse CARM1 in complex with ligands
To Be Published
|
|
2ZYS
 
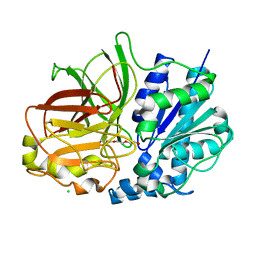 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
1AJJ
 
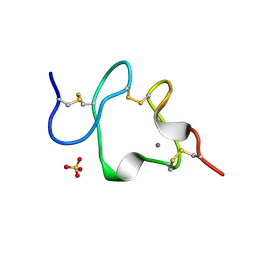 | | LDL RECEPTOR LIGAND-BINDING MODULE 5, CALCIUM-COORDINATING | | Descriptor: | CALCIUM ION, LOW-DENSITY LIPOPROTEIN RECEPTOR, SULFATE ION | | Authors: | Fass, D, Blacklow, S.C, Kim, P.S, Berger, J.M. | | Deposit date: | 1997-05-04 | | Release date: | 1997-07-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular basis of familial hypercholesterolaemia from structure of LDL receptor module.
Nature, 388, 1997
|
|
2MVV
 
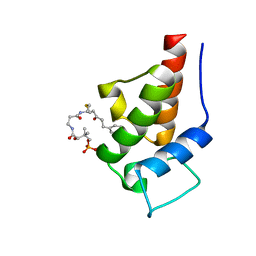 | | Solution Structure of the 5-phenyl-3-oxo-pentyl Actinorhodin Acyl Carrier Protein from Streptomyces coelicolor | | Descriptor: | Actinorhodin polyketide synthase acyl carrier protein, N~3~-{(2S)-4-[(dihydroxyphosphanyl)oxy]-2-hydroxy-3,3-dimethylbutanoyl}-N-{2-[(3-oxo-5-phenylpentyl)sulfanyl]ethyl}-beta-alaninamide | | Authors: | Dong, X, Bailey, C, Williams, C, Crosby, J, Simpson, T.J, Willis, C.L, Crump, M.P. | | Deposit date: | 2014-10-15 | | Release date: | 2015-10-21 | | Last modified: | 2024-10-09 | | Method: | SOLUTION NMR | | Cite: | ACP-ligand recognition: Selection of derivatized aromatic biosynthetic intermediates
To be Published
|
|
1QHK
 
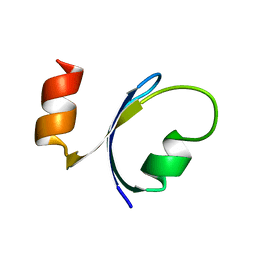 | |
5EYZ
 
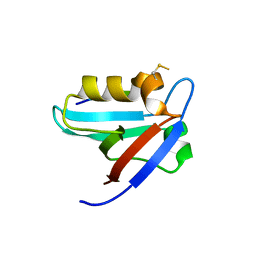 | | CRYSTAL STRUCTURE OF THE PTPN4 PDZ DOMAIN COMPLEXED WITH THE TAILORED PEPTIDE CYTO8-RETEV | | Descriptor: | CHLORIDE ION, CYTO8-RETEV, Tyrosine-protein phosphatase non-receptor type 4 | | Authors: | Maisonneuve, P, Vaney, M.C, Babault, B, Caillet-Saguy, C, Lafon, M, Delepierre, M, Cordier, F, Wolff, N. | | Deposit date: | 2015-11-26 | | Release date: | 2016-06-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Molecular Basis of the Interaction of the Human Protein Tyrosine Phosphatase Non-receptor Type 4 (PTPN4) with the Mitogen-activated Protein Kinase p38 gamma.
J.Biol.Chem., 291, 2016
|
|
2ZX5
 
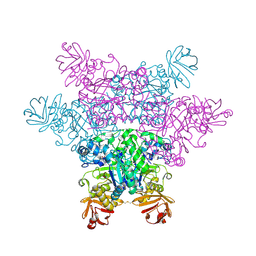 | | alpha-L-fucosidase complexed with inhibitor, F10 | | Descriptor: | 3-(1H-indol-3-yl)-N-{[(2R,3R,4R,5R,6S)-3,4,5-trihydroxy-6-methylpiperidin-2-yl]methyl}propanamide, Alpha-L-fucosidase, putative | | Authors: | Wu, H.-J, Ko, T.-P, Ho, C.-W, Lin, C.-H, Wang, A.H.-J. | | Deposit date: | 2008-12-19 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis of alpha-fucosidase inhibition by iminocyclitols with K(i) values in the micro- to picomolar range.
Angew.Chem.Int.Ed.Engl., 49, 2010
|
|
2MP2
 
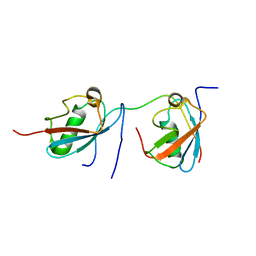 | | Solution structure of SUMO dimer in complex with SIM2-3 from RNF4 | | Descriptor: | E3 ubiquitin-protein ligase RNF4, Small ubiquitin-related modifier 3 | | Authors: | Xu, Y, Plechanovov, A, Simpson, P, Marchant, J, Leidecker, O, Sebastian, K, Hay, R.T, Matthews, S.J. | | Deposit date: | 2014-05-09 | | Release date: | 2014-07-02 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into SUMO chain recognition and manipulation by the ubiquitin ligase RNF4.
Nat Commun, 5, 2014
|
|
