1BTL
 
 | | CRYSTAL STRUCTURE OF ESCHERICHIA COLI TEM1 BETA-LACTAMASE AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | BETA-LACTAMASE TEM1, SULFATE ION | | Authors: | Jelsch, C, Mourey, L, Masson, J.M, Samama, J.P. | | Deposit date: | 1993-11-01 | | Release date: | 1995-01-26 | | Last modified: | 2019-08-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of Escherichia coli TEM1 beta-lactamase at 1.8 A resolution.
Proteins, 16, 1993
|
|
6ST3
 
 | | HIF prolyl hydroxylase 2 (PHD2/ EGLN1) in complex with 4-hydroxy-N-(4-phenoxybenzyl)-2-(1H-pyrazol-1-yl)pyrimidine-5-carboxamide | | Descriptor: | 4-oxidanyl-~{N}-[(4-phenoxyphenyl)methyl]-2-pyrazol-1-yl-pyrimidine-5-carboxamide, Egl nine homolog 1, FORMIC ACID, ... | | Authors: | Chowdhury, R, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.426 Å) | | Cite: | Structure-Activity Relationship and Crystallographic Studies on 4-Hydroxypyrimidine HIF Prolyl Hydroxylase Domain Inhibitors.
Chemmedchem, 15, 2020
|
|
4XX4
 
 | |
5FZ4
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment (3R)-1-[(3-phenyl-1,2,4-oxadiazol-5-yl)methyl]pyrrolidin-3-ol (N10057a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | (3S)-1-[(3-phenyl-1,2,4-oxadiazol-5-yl)methyl]pyrrolidin-3-ol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Pearce, N, Talon, R, Collins, P, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N10057A
To be Published
|
|
5FZC
 
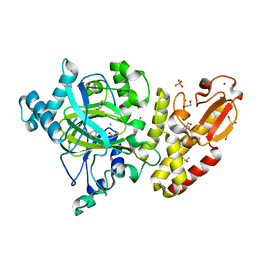 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 4,5-dihydronaphtho(1,2-b)thiophene-2- carboxylicacid (N11181a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment 4,5-Dihydronaphtho(1,2-B)Thiophene-2-Carboxylicacid (N11181A) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5FYW
 
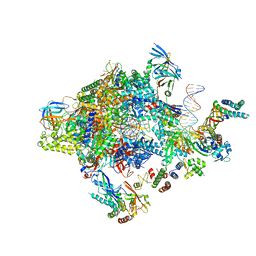 | | Transcription initiation complex structures elucidate DNA opening (OC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.35 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
4Y2H
 
 | |
5FZL
 
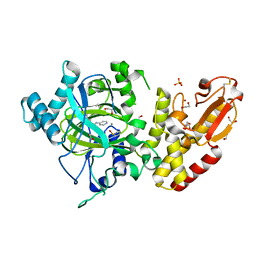 | | Crystal structure of the catalytic domain of human JARID1B in complex with 3D fragment 3-methyl-N-pyridin-4-yl-1,2-oxazole-5-carboxamide (N09954a) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 3-methyl-N-(pyridin-4-yl)-1,2-oxazole-5-carboxamide, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with 3D Fragment 3-Methyl-N-Pyridin-4-Yl-1,2-Oxazole-5-Carboxamide (N09954A) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
5FMK
 
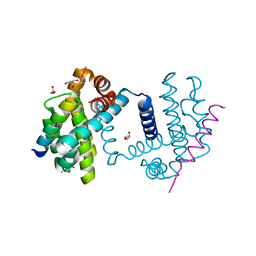 | | Bcl-xL with Bak BH3 complex | | Descriptor: | BCL-2 HOMOLOGOUS ANTAGONIST/KILLER, BCL-XL, GLYCEROL | | Authors: | Fairlie, W.D, Lee, E.F, Smith, B.J, Czabotar, P.E, Colman, P.M. | | Deposit date: | 2015-11-06 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.731 Å) | | Cite: | Physiological Restraint of Bak by Bcl-Xl is Essential for Cell Survival.
Genes Dev., 30, 2016
|
|
5FD3
 
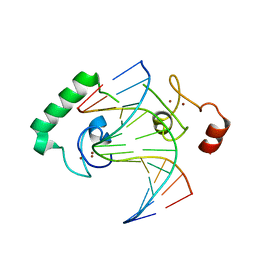 | | Structure of Lin54 tesmin domain bound to DNA | | Descriptor: | DNA (5'-D(*CP*AP*GP*TP*TP*TP*CP*AP*AP*AP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*TP*TP*TP*GP*AP*AP*AP*CP*T)-3'), Protein lin-54 homolog, ... | | Authors: | Marceau, A.H, Felthousen, J.G, Goetsch, P.D, Lee, H, Tripathi, S.M, Strome, S, Litovchick, L, Rubin, S.M. | | Deposit date: | 2015-12-15 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural basis for LIN54 recognition of CHR elements in cell cycle-regulated promoters.
Nat Commun, 7, 2016
|
|
5FO9
 
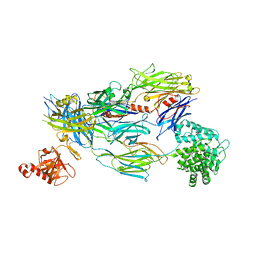 | | Crystal Structure of Human Complement C3b in Complex with CR1 (CCP15- 17) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COMPLEMENT C3 BETA CHAIN, COMPLEMENT C3B ALPHA' CHAIN, ... | | Authors: | Forneris, F, Wu, J, Xue, X, Gros, P. | | Deposit date: | 2015-11-18 | | Release date: | 2016-04-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Regulators of Complement Activity Mediate Inhibitory Mechanisms Through a Common C3B-Binding Mode.
Embo J., 35, 2016
|
|
6MJW
 
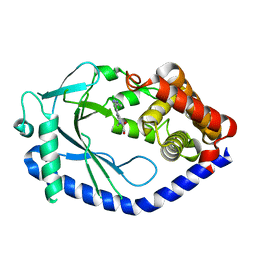 | | human cGAS catalytic domain bound with the inhibitor G150 | | Descriptor: | 1-[9-(6-aminopyridin-3-yl)-6,7-dichloro-1,3,4,5-tetrahydro-2H-pyrido[4,3-b]indol-2-yl]-2-hydroxyethan-1-one, Cyclic GMP-AMP synthase, ZINC ION | | Authors: | Lama, L, Adura, C, Xie, W, Tomita, D, Kamei, T, Kuryavyi, V, Gogakos, T, Steinberg, J.I, Miller, M, Ramos-Espiritu, L, Asano, Y, Hashizume, S, Aida, J, Imaeda, T, Okamoto, R, Jennings, A.J, Michinom, M, Kuroita, T, Stamford, A, Gao, P, Meinke, P, Glickman, J.F, Patel, D.J, Tuschl, T. | | Deposit date: | 2018-09-23 | | Release date: | 2019-05-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Development of human cGAS-specific small-molecule inhibitors for repression of dsDNA-triggered interferon expression.
Nat Commun, 10, 2019
|
|
5FQU
 
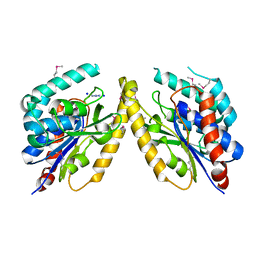 | | Orthorhombic crystal structure of of PlpD (selenomethionine derivative) | | Descriptor: | AZIDE ION, PATATIN-LIKE PROTEIN, PLPD, ... | | Authors: | Vinicius da Mata Madeira, P, Zouhir, S, Basso, P, Neves, D, Laubier, A, Salacha, R, Bleves, S, Faudry, E, Contreras-Martel, C, Dessen, A. | | Deposit date: | 2015-12-14 | | Release date: | 2016-04-06 | | Last modified: | 2017-09-27 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural Basis of Lipid Targeting and Destruction by the Type V Secretion System of Pseudomonas Aeruginosa.
J.Mol.Biol., 428, 2016
|
|
8I8P
 
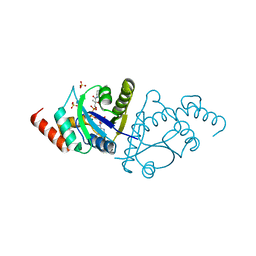 | | Crystal structure of the complex of phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephosphocoenzyme-A at 2.19 A resolution. | | Descriptor: | CHLORIDE ION, DEPHOSPHO COENZYME A, MAGNESIUM ION, ... | | Authors: | Ahmad, N, Viswanathan, V, Gupta, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2023-02-04 | | Release date: | 2023-04-12 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal structure of the complex of phosphopantetheine adenylyltransferase from Acinetobacter baumannii with Dephosphocoenzyme-A at 2.19 A resolution.
To Be Published
|
|
5MBQ
 
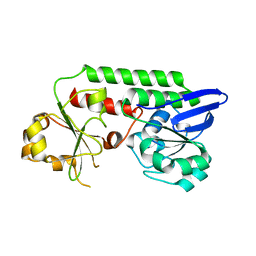 | | CeuE (H227A variant) a periplasmic protein from Campylobacter jejuni | | Descriptor: | Enterochelin uptake periplasmic binding protein | | Authors: | Wilde, E.J, Blagova, E.V, Hughes, A, Raines, D.J, Moroz, O.V, Turkenburg, J.P, Duhme-Klair, A.-K, Wilson, K.S. | | Deposit date: | 2016-11-08 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Interactions of the periplasmic binding protein CeuE with Fe(III) n-LICAM(4-) siderophore analogues of varied linker length.
Sci Rep, 7, 2017
|
|
7PHD
 
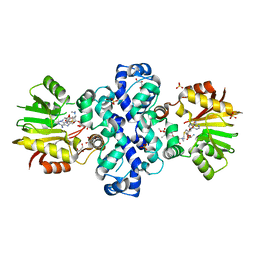 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with a region from 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein, GLYCEROL, S-ADENOSYLMETHIONINE, ... | | Authors: | Grocholski, T, Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
5FZ5
 
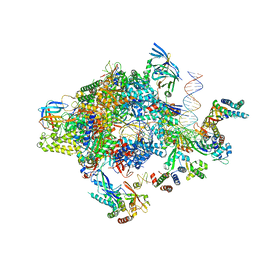 | | Transcription initiation complex structures elucidate DNA opening (CC) | | Descriptor: | DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB1, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB11, DNA-DIRECTED RNA POLYMERASE II SUBUNIT RPB2, ... | | Authors: | Plaschka, C, Hantsche, M, Dienemann, C, Burzinski, C, Plitzko, J, Cramer, P. | | Deposit date: | 2016-03-10 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (8.8 Å) | | Cite: | Transcription Initiation Complex Structures Elucidate DNA Opening
Nature, 533, 2016
|
|
7PHE
 
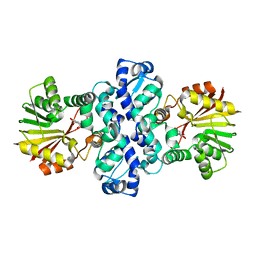 | | Chimeric carminomycin-4-O-methyltransferase (DnrK) with regions from 10-hydroxylase RdmB and 10-decarboxylase TamK | | Descriptor: | Carminomycin 4-O-methyltransferase DnrK,Methyltransferase domain-containing protein,Aclacinomycin 10-hydroxylase RdmB, methyl (1R,2R,4S)-2-ethyl-2,4,5,7-tetrahydroxy-6,11-dioxo-1,2,3,4,6,11-hexahydrotetracene-1-carboxylate | | Authors: | Dinis, P, MetsaKetela, M. | | Deposit date: | 2021-08-17 | | Release date: | 2022-09-07 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Evolution-inspired engineering of anthracycline methyltransferases.
Pnas Nexus, 2, 2023
|
|
7UCE
 
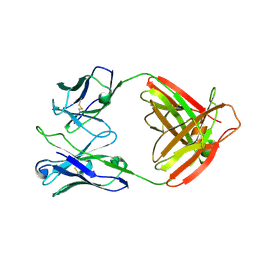 | |
3IVK
 
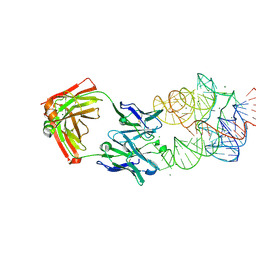 | | Crystal Structure of the Catalytic Core of an RNA Polymerase Ribozyme Complexed with an Antigen Binding Antibody Fragment | | Descriptor: | CADMIUM ION, CHLORIDE ION, Fab heavy chain, ... | | Authors: | Koldobskaya, Y, Duguid, E.M, Shechner, D.M, Koide, S, Kossiakoff, A.A, Bartel, D.P, Piccirilli, J.A. | | Deposit date: | 2009-09-01 | | Release date: | 2010-03-02 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal structure of the catalytic core of an RNA-polymerase ribozyme.
Science, 326, 2009
|
|
5MFD
 
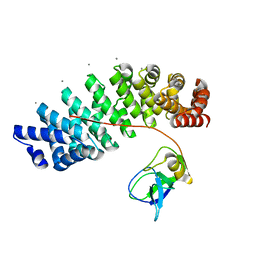 | | Designed armadillo repeat protein YIIIM''6AII in complex with pD_(KR)5 | | Descriptor: | CALCIUM ION, Capsid decoration protein,pD_(KR)5, YIIIM''6AII | | Authors: | Hansen, S, Kiefer, J, Madhurantakam, C, Mittl, P, Plueckthun, A. | | Deposit date: | 2016-11-18 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of designed armadillo repeat proteins binding to peptides fused to globular domains.
Protein Sci., 26, 2017
|
|
5FH8
 
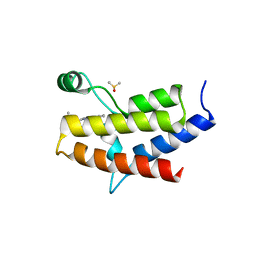 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 28 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-(2-ethylbutyl)-4~{H}-pyrrolo[1,2-a]quinazolin-5-one, DIMETHYL SULFOXIDE, ... | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Sorrell, F.J, Krojer, T, Picaud, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
6FCM
 
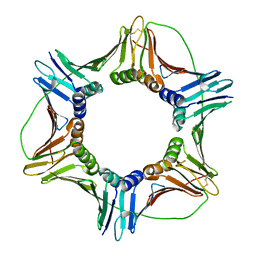 | | Crystal structure of human PCNA | | Descriptor: | Proliferating cell nuclear antigen | | Authors: | Housset, D, Frachet, P. | | Deposit date: | 2017-12-20 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Cytosolic PCNA interacts with p47phox and controls NADPH oxidase NOX2 activation in neutrophils.
J.Exp.Med., 216, 2019
|
|
8A5V
 
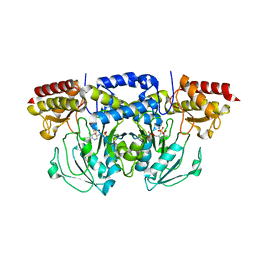 | | Crystal structure of the human phosposerine aminotransferase (PSAT) | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase, SULFATE ION | | Authors: | Costanzi, E, Demitri, N, Ullah, R, Marchesan, F, Peracchi, A, Zangelmi, E, Storici, P, Campanini, B. | | Deposit date: | 2022-06-16 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | L-serine biosynthesis in the human central nervous system: Structure and function of phosphoserine aminotransferase.
Protein Sci., 32, 2023
|
|
5FZ9
 
 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment thieno(3,2-b)thiophene-5-carboxylic acid (N06263b) (ligand modelled based on PANDDA event map, SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Kupinska, K, Szykowska, A, Pearce, N, Talon, R, Collins, P, Gileadi, C, Strain-Damerell, C, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-12 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with Maybridge Fragment Thieno(3,2-B)Thiophene -5-Carboxylic Acid (N06263B) (Ligand Modelled Based on Pandda Event Map, Sgc - Diamond I04-1 Fragment Screening)
To be Published
|
|
