3HK6
 
 | | Crystal structure of murine thrombin mutant W215A/E217A (two molecules in the asymmetric unit) | | Descriptor: | Thrombin heavy chain, Thrombin light chain | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-22 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
3HKI
 
 | | Crystal structure of murine thrombin mutant W215A/E217A in complex with the extracellular fragment of human PAR1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Proteinase-activated receptor 1, Thrombin heavy chain, ... | | Authors: | Gandhi, P.S, Page, M.J, Chen, Z, Bush-Pelc, L, Di Cera, E. | | Deposit date: | 2009-05-23 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanism of the Anticoagulant Activity of Thrombin Mutant W215A/E217A.
J.Biol.Chem., 284, 2009
|
|
5QJC
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z755044716 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
2H1K
 
 | |
5QJN
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1439422127 | | Descriptor: | (2S,3R)-2,3-dimethyl-4-(3-methyl-1,2,4-thiadiazol-5-yl)morpholine, 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-11-12 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QK4
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z44590919 | | Descriptor: | 1,2-ETHANEDIOL, 4-acetyl-N-ethylpiperazine-1-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4DMB
 
 | | X-ray structure of human hepatitus C virus NS5A-transactivated protein 2 at the resolution 1.9A, Northeast Structural Genomics Consortium (NESG) Target HR6723 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, HD domain-containing protein 2, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2012-02-07 | | Release date: | 2012-04-04 | | Last modified: | 2012-04-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR6723
To be Published
|
|
4DOS
 
 | | Human Nuclear Receptor Liver Receptor Homologue-1, LRH-1, Bound to DLPC and a Fragment of TIF-2 | | Descriptor: | DIUNDECYL PHOSPHATIDYL CHOLINE, Nuclear receptor coactivator 2, Nuclear receptor subfamily 5 group A member 2, ... | | Authors: | Musille, P.M, Ortlund, E.A. | | Deposit date: | 2012-02-10 | | Release date: | 2012-04-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation.
Nat.Struct.Mol.Biol., 19, 2012
|
|
5QJH
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z375990520 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QJW
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z198194396 | | Descriptor: | 1,2-ETHANEDIOL, 4-(furan-2-carbonyl)piperazine-1-carboxamide, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1DX5
 
 | | Crystal structure of the thrombin-thrombomodulin complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, FORMIC ACID, ... | | Authors: | Fuentes-Prior, P, Iwanaga, Y, Huber, R, Pagila, R, Rumennik, G, Seto, M, Morser, J, Light, D.R, Bode, W. | | Deposit date: | 1999-12-20 | | Release date: | 2000-04-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Anticoagulant Activity of the Thrombin-Thrombomodulin Complex
Nature, 404, 2000
|
|
5QK0
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1270312110 | | Descriptor: | 1,2-ETHANEDIOL, 2-(3,5-dimethyl-1H-pyrazol-4-yl)aniline, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1POS
 
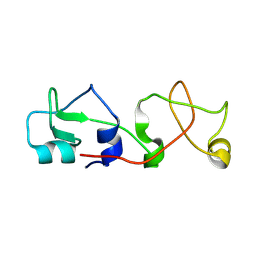 | | CRYSTAL STRUCTURE OF A NOVEL DISULFIDE-LINKED "TREFOIL" MOTIF FOUND IN A LARGE FAMILY OF PUTATIVE GROWTH FACTORS | | Descriptor: | PORCINE PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | De, A, Brown, D, Gorman, M, Carr, M, Sanderson, M.R, Freemont, P.S. | | Deposit date: | 1993-10-08 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a disulfide-linked "trefoil" motif found in a large family of putative growth factors.
Proc.Natl.Acad.Sci.USA, 91, 1994
|
|
3QFT
 
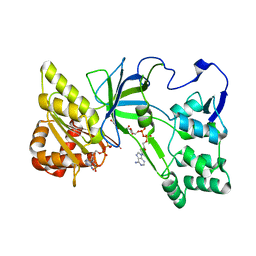 | | Crystal Structure of NADPH-Cytochrome P450 Reductase (FAD/NADPH domain and R457H Mutant) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH--cytochrome P450 reductase | | Authors: | Xia, C, Marohnic, C, Panda, S.P, Masters, B.S, Kim, J.-J.P. | | Deposit date: | 2011-01-22 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for human NADPH-cytochrome P450 oxidoreductase deficiency.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2AZX
 
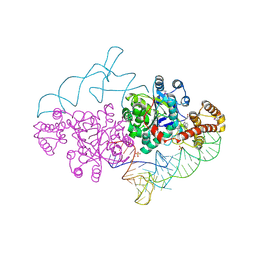 | | Charged and uncharged tRNAs adopt distinct conformations when complexed with human tryptophanyl-tRNA synthetase | | Descriptor: | 72-MER, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Yang, X.L, Otero, F.J, Ewalt, K.L, Liu, J, Swairjo, M.A, Kohrer, C, RajBhandary, U.L, Skene, R.J, McRee, D.E, Schimmel, P. | | Deposit date: | 2005-09-12 | | Release date: | 2006-08-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Two conformations of a crystalline human tRNA synthetase-tRNA complex: implications for protein synthesis.
Embo J., 25, 2006
|
|
5QJ5
 
 | | PanDDA analysis group deposition -- Crystal Structure of NUDT5 in complex with Z44592329 | | Descriptor: | 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, CHLORIDE ION, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | PanDDA analysis group deposition
To Be Published
|
|
1JE4
 
 | |
5QJK
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z373221060 | | Descriptor: | 1,2-ETHANEDIOL, 6-(azetidin-1-yl)-9H-purine, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4RCH
 
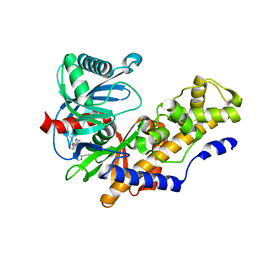 | | Discovery of 2-Pyridyl Ureas as Glucokinase Activators | | Descriptor: | 1-{3-[(2-ethylpyridin-3-yl)oxy]-5-(pyridin-2-ylsulfanyl)pyridin-2-yl}-3-methylurea, Glucokinase, alpha-D-glucopyranose | | Authors: | Voegtli, W, Vigers, G.P.A. | | Deposit date: | 2014-09-16 | | Release date: | 2014-10-08 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of 2-pyridylureas as glucokinase activators.
J.Med.Chem., 57, 2014
|
|
3BJU
 
 | | Crystal Structure of tetrameric form of human lysyl-tRNA synthetase | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, LYSINE, ... | | Authors: | Guo, M, Yang, X.L, Schimmel, P. | | Deposit date: | 2007-12-04 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal structure of tetrameric form of human lysyl-tRNA synthetase: Implications for multisynthetase complex formation
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
5QJZ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT5 in complex with Z1696822287 | | Descriptor: | (1R)-1-[4-(pyrimidin-5-yl)phenyl]ethan-1-amine, 1,2-ETHANEDIOL, ADP-sugar pyrophosphatase, ... | | Authors: | Dubianok, Y, Collins, P, Krojer, T, Wright, N, Strain-Damerell, C, Burgess-Brown, N, Bountra, C, Arrowsmith, C.H, Edwards, A, Huber, K, von Delft, F. | | Deposit date: | 2018-10-31 | | Release date: | 2018-12-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
1DV9
 
 | | STRUCTURAL CHANGES ACCOMPANYING PH-INDUCED DISSOCIATION OF THE B-LACTOGLOBULIN DIMER | | Descriptor: | BETA-LACTOGLOBULIN | | Authors: | Uhrinova, S, Smith, M.H, Jameson, G.B, Uhrin, D, Sawyer, L, Barlow, P.N. | | Deposit date: | 2000-01-20 | | Release date: | 2000-02-09 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Structural changes accompanying pH-induced dissociation of the beta-lactoglobulin dimer.
Biochemistry, 39, 2000
|
|
3IZQ
 
 | | Structure of the Dom34-Hbs1-GDPNP complex bound to a translating ribosome | | Descriptor: | Elongation factor 1 alpha-like protein, Protein DOM34 | | Authors: | Becker, T, Armache, J.-P, Jarasch, A, Anger, A.M, Villa, E, Sieber, H, Abdel Motaal, B, Mielke, T, Berninghausen, O, Beckmann, R. | | Deposit date: | 2010-11-30 | | Release date: | 2011-06-01 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1YBY
 
 | | Conserved hypothetical protein Cth-95 from Clostridium thermocellum | | Descriptor: | Translation elongation factor P, UNKNOWN ATOM OR ION | | Authors: | Zhao, M, Zhou, W, Chang, J, Habel, J, Kataeva, I, Xu, H, Chen, L, Lee, D, Nguyen, J, Chang, S.-H, Horanyi, P, Florence, Q, Tempel, W, Lin, D, Zhang, H, Ljundahl, L, Liu, Z.-J, Rose, J, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-21 | | Release date: | 2005-02-01 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Conserved hypothetical protein Cth-95 from Clostridium thermocellum
To be published
|
|
2HA4
 
 | | Crystal structure of mutant S203A of mouse acetylcholinesterase complexed with acetylcholine | | Descriptor: | ACETATE ION, ACETYLCHOLINE, Acetylcholinesterase, ... | | Authors: | Bourne, Y, Radic, Z, Sulzenbacher, G, Kim, E, Taylor, P, Marchot, P. | | Deposit date: | 2006-06-12 | | Release date: | 2006-07-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Substrate and product trafficking through the active center gorge of acetylcholinesterase analyzed by crystallography and equilibrium binding
J.Biol.Chem., 281, 2006
|
|
