9ENP
 
 | | HSV-1 DNA polymerase-processivity factor complex in exonuclease state with 1-bp DNA mismatch | | Descriptor: | CALCIUM ION, DNA (46-MER), DNA (67-MER), ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2024-03-13 | | Release date: | 2024-05-29 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.12 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
8ZN1
 
 | | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 3.00 A resolution
To Be Published
|
|
9BKM
 
 | | DHODH in complex with Ligand 10 | | Descriptor: | (2M,6P)-2-(2-chloro-6-fluorophenyl)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-4-(propan-2-yl)isoquinolin-1(2H)-one, ACETATE ION, Dihydroorotate dehydrogenase (quinone), ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 67, 2024
|
|
9F3E
 
 | |
4W8K
 
 | | Crystal structure of a putative Cas1 enzyme from Vibrio phage ICP1 | | Descriptor: | Cas1 protein, POTASSIUM ION | | Authors: | Stogios, P.J, Wawrzak, Z, Onopriyeno, O, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | To be published
To Be Published
|
|
9BKN
 
 | | DHODH in complex with Ligand 16 | | Descriptor: | (2P,6P)-6-[4-ethyl-3-(hydroxymethyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-1-yl]-7-fluoro-2-(2-methylphenyl)-4-(propan-2-yl)isoquinolin-1(2H)-one, ACETATE ION, CHLORIDE ION, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Discovery of JNJ-74856665: A Novel Isoquinolinone DHODH Inhibitor for the Treatment of AML.
J.Med.Chem., 67, 2024
|
|
9IIM
 
 | | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution. | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Jeyakanthan, J, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-06-20 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structure of the complex of erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii with nicotinamide adenine dinucleotide at 2.74 A resolution.
To Be Published
|
|
8ZN4
 
 | | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3,6,9,12,15,18,21,24,27-NONAOXANONACOSANE-1,29-DIOL, ... | | Authors: | Viswanathan, V, Kumari, A, Singh, A, Kumar, A, Sharma, P, Chopra, S, Sharma, S, Raje, C.I, Singh, T.P. | | Deposit date: | 2024-05-25 | | Release date: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Poly(ethylene glycol) stabilized erythrose-4-phosphate dehydrogenase from Acinetobacter baumannii at 2.30 A resolution
To Be Published
|
|
9B37
 
 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9B36
 
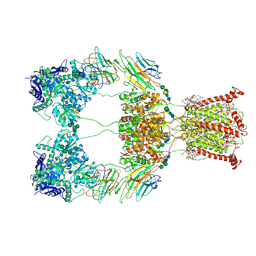 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to two concanavalin A dimers. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.29 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
5HOR
 
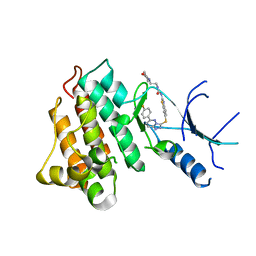 | | Crystal structure of c-Met-M1250T in complex with SAR125844. | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Houtmann, J, Marquette, J.-P. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
5HLW
 
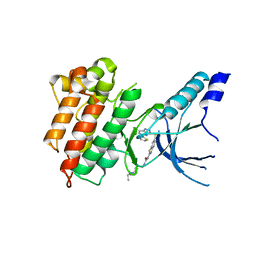 | | Crystal structure of c-Met mutant Y1230H in complex with compound 14 | | Descriptor: | 1-[2-(1-ethylpiperidin-4-yl)ethyl]-3-(6-{[6-(thiophen-2-yl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)urea, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Pouzieux, S, Marquette, J.P, Houtmann, J. | | Deposit date: | 2016-01-15 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
4WED
 
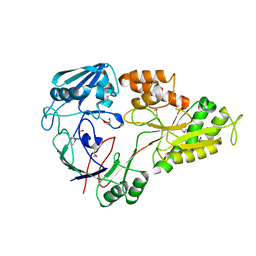 | | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti | | Descriptor: | ABC transporter, periplasmic solute-binding protein, FORMIC ACID, ... | | Authors: | Shabalin, I.G, Otwinowski, Z, Bacal, P, Cymborowski, M.T, Handing, K.B, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-09 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of ABC transporter substrate-binding protein from Sinorhizobium meliloti
to be published
|
|
19HC
 
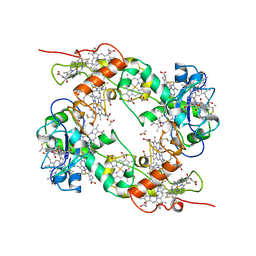 | | NINE-HAEM CYTOCHROME C FROM DESULFOVIBRIO DESULFURICANS ATCC 27774 | | Descriptor: | ACETATE ION, PROTEIN (NINE-HAEM CYTOCHROME C), PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Matias, P.M, Coelho, R, Pereira, I.A.C, Coelho, A.V, Thompson, A.W, Sieker, L, Gall, J.L, Carrondo, M.A. | | Deposit date: | 1998-12-01 | | Release date: | 1999-12-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The primary and three-dimensional structures of a nine-haem cytochrome c from Desulfovibrio desulfuricans ATCC 27774 reveal a new member of the Hmc family.
Structure Fold.Des., 7, 1999
|
|
5HOA
 
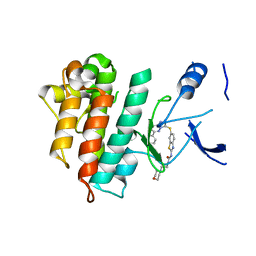 | | Crystal structure of c-Met L1195V in complex with SAR125844 | | Descriptor: | 1-(6-{[6-(4-fluorophenyl)[1,2,4]triazolo[4,3-b]pyridazin-3-yl]sulfanyl}-1,3-benzothiazol-2-yl)-3-[2-(morpholin-4-yl)ethyl]urea, Hepatocyte growth factor receptor | | Authors: | Vallee, F, Marquette, J.-P. | | Deposit date: | 2016-01-19 | | Release date: | 2016-11-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Discovery and Pharmacokinetic and Pharmacological Properties of the Potent and Selective MET Kinase Inhibitor 1-{6-[6-(4-Fluorophenyl)-[1,2,4]triazolo[4,3-b]pyridazin-3-ylsulfanyl]benzothiazol-2-yl}-3-(2-morpholin-4-ylethyl)urea (SAR125844).
J.Med.Chem., 59, 2016
|
|
129D
 
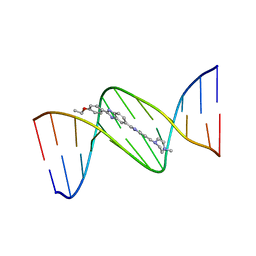 | | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') COMPLEXED WITH HOECHST 33342 | | Descriptor: | 2'-(4-ETHOXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
127D
 
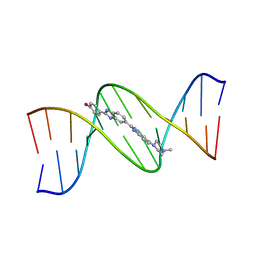 | | DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') COMPLEXED WITH HOECHST 33258 | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1993-06-30 | | Release date: | 1994-01-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Conformation of B-DNA containing O6-ethyl-G-C base pairs stabilized by minor groove binding drugs: molecular structure of d(CGC[e6G]AATTCGCG complexed with Hoechst 33258 or Hoechst 33342.
EMBO J., 11, 1992
|
|
129L
 
 | |
130L
 
 | |
131L
 
 | |
2E6W
 
 | | Solution structure and calcium binding properties of EF-hands 3 and 4 of calsenilin | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Yu, L, Sun, C, Mendoza, R, Hebert, E, Pereda-Lopez, A, Hajduk, P.J, Olejniczak, E.T. | | Deposit date: | 2007-01-05 | | Release date: | 2007-11-27 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure and calcium-binding properties of EF-hands 3 and 4 of calsenilin.
Protein Sci., 16, 2007
|
|
1AI0
 
 | | R6 HUMAN INSULIN HEXAMER (NON-SYMMETRIC), NMR, 10 STRUCTURES | | Descriptor: | PHENOL, R6 INSULIN HEXAMER, ZINC ION | | Authors: | Chang, X, Jorgensen, A.M.M, Bardrum, P, Led, J.J. | | Deposit date: | 1997-04-30 | | Release date: | 1997-11-12 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structures of the R6 human insulin hexamer.
Biochemistry, 36, 1997
|
|
2EBJ
 
 | | Crystal structure of pyrrolidone carboxyl peptidase from Thermus thermophilus | | Descriptor: | Pyrrolidone carboxyl peptidase | | Authors: | Kumarevel, T.S, Karthe, P, Agari, Y, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-02-08 | | Release date: | 2007-08-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of pyrrolidone carboxyl peptidase from Thermus thermophilus
To be Published
|
|
4WA0
 
 | |
156D
 
 | |
