5PD3
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 59) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | 著者 | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2017-02-03 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PDQ
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 83) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | 著者 | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2017-02-03 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (2.26 Å) | | 主引用文献 | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4APB
 
 | | Crystal structure of Mycobacterium tuberculosis fumarase (Rv1098c) S318C in complex with fumarate | | 分子名称: | CALCIUM ION, FUMARATE HYDRATASE CLASS II, FUMARIC ACID | | 著者 | Bellinzoni, M, Haouz, A, Mechaly, A.E, Alzari, P.M. | | 登録日 | 2012-03-31 | | 公開日 | 2012-04-25 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Conformational Changes Upon Ligand Binding in the Essential Class II Fumarase Rv1098C from Mycobacterium Tuberculosis.
FEBS Lett., 586, 2012
|
|
5PE5
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 98) | | 分子名称: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | 著者 | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2017-02-03 | | 公開日 | 2017-03-22 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
2X70
 
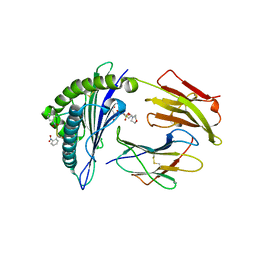 | | Crystal structure of MHC CLass I HLA-A2.1 bound to a photocleavable peptide | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BETA-2-MICROGLOBULIN, GLYCEROL, ... | | 著者 | Celie, P.H.N, Toebes, M, Rodenko, B, Ovaa, H, Perrakis, A, Schumacher, T.N.M. | | 登録日 | 2010-02-22 | | 公開日 | 2010-12-08 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Uv-Induced Ligand Exchange in Mhc Class I Protein Crystals.
J.Am.Chem.Soc., 131, 2009
|
|
4G92
 
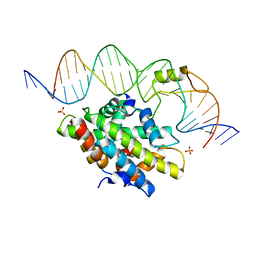 | | CCAAT-binding complex from Aspergillus nidulans with DNA | | 分子名称: | DNA, HAPB protein, HapE, ... | | 著者 | Huber, E.M, Scharf, D.H, Hortschansky, P, Groll, M, Brakhage, A.A. | | 登録日 | 2012-07-23 | | 公開日 | 2012-10-31 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | DNA Minor Groove Sensing and Widening by the CCAAT-Binding Complex.
Structure, 20, 2012
|
|
3DOK
 
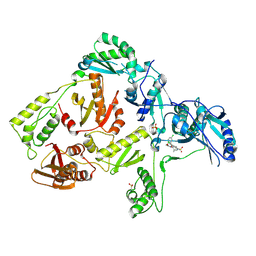 | | Crystal structure of K103N mutant HIV-1 reverse transcriptase in complex with GW678248. | | 分子名称: | 2-{4-chloro-2-[(3-chloro-5-cyanophenyl)carbonyl]phenoxy}-N-(2-methyl-4-sulfamoylphenyl)acetamide, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | 著者 | Chamberlain, P.P, Ren, J, Stammers, D.K. | | 登録日 | 2008-07-04 | | 公開日 | 2008-08-12 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural basis for the improved drug resistance profile of new generation benzophenone non-nucleoside HIV-1 reverse transcriptase inhibitors.
J.Med.Chem., 51, 2008
|
|
1KRC
 
 | |
1KRB
 
 | |
1E32
 
 | | Structure of the N-Terminal domain and the D1 AAA domain of membrane fusion ATPase p97 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, P97 | | 著者 | Zhang, X, Shaw, A, Bates, P.A, Gorman, M.A, Kondo, H, Dokurno, P, Leonard M, G, Sternberg, J.E, Freemont, P.S. | | 登録日 | 2000-06-05 | | 公開日 | 2001-05-31 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of the Aaa ATPase P97
Mol.Cell, 6, 2000
|
|
4A30
 
 | | CRYSTAL STRUCTURE OF LEISHMANIA MAJOR N-MYRISTOYLTRANSFERASE (NMT) WITH BOUND MYRISTOYL-COA AND A PYRAZOLE SULPHONAMIDE LIGAND | | 分子名称: | 4-BROMO-2,6-DICHLORO-N-(1,3,5-TRIMETHYL-1H-PYRAZOL-4-YL)BENZENESULFONAMIDE, CHLORIDE ION, GLYCEROL, ... | | 著者 | Robinson, D.A, Brand, S, Fairlamb, A.H, Ferguson, M.A.J, Frearson, J.A, Wyatt, P.G. | | 登録日 | 2011-09-29 | | 公開日 | 2011-12-21 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Discovery of a novel class of orally active trypanocidal N-myristoyltransferase inhibitors.
J. Med. Chem., 55, 2012
|
|
1PIN
 
 | | PIN1 PEPTIDYL-PROLYL CIS-TRANS ISOMERASE FROM HOMO SAPIENS | | 分子名称: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, ALANINE, PEPTIDYL-PROLYL CIS-TRANS ISOMERASE, ... | | 著者 | Noel, J.P, Ranganathan, R, Hunter, T. | | 登録日 | 1998-06-21 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.35 Å) | | 主引用文献 | Structural and functional analysis of the mitotic rotamase Pin1 suggests substrate recognition is phosphorylation dependent.
Cell(Cambridge,Mass.), 89, 1997
|
|
2PTQ
 
 | |
4MXE
 
 | | Human ESCO1 (Eco1/Ctf7 ortholog), acetyltransferase domain in complex with acetyl-CoA | | 分子名称: | ACETYL COENZYME *A, N-acetyltransferase ESCO1 | | 著者 | Karlberg, T, Wisniewska, M, Thorsell, A.G, Kouznetsova, E, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, I, Kallas, A, Kraulis, P, Kotenyova, T, Moche, M, Nielsen, T.K, Nordlund, P, Nyman, T, Persson, C, Schutz, P, Svensson, L, Tresaugues, L, Van Den Berg, S, Wahlberg, E, Weigelt, J, Welin, M, Schuler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2013-09-26 | | 公開日 | 2015-04-08 | | 最終更新日 | 2016-05-18 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Sister Chromatid Cohesion Establishment Factor ESCO1 Operates by Substrate-Assisted Catalysis.
Structure, 24, 2016
|
|
4DDZ
 
 | | Crystal structure of glucosyl-3-phosphoglycerate synthase from Mycobacterium tuberculosis | | 分子名称: | GLUCOSYL-3-PHOSPHOGLYCERATE SYNTHASE (GpgS), GLYCEROL | | 著者 | Albesa-Jove, D, Urresti, S, Gest, P.M, van der Woerd, M, Jackson, M, Guerin, M.E. | | 登録日 | 2012-01-19 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Mechanistic insights into the retaining glucosyl-3-phosphoglycerate synthase from mycobacteria.
J.Biol.Chem., 287, 2012
|
|
4MYO
 
 | | Crystal structure of streptogramin group A antibiotic acetyltransferase VatA from Staphylococcus aureus | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, SULFATE ION, ... | | 著者 | Stogios, P.J, Minasov, G, Dong, A, Evdokimova, E, Yim, V, Krishnamoorthy, M, Di Leo, R, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2013-09-27 | | 公開日 | 2013-10-16 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.696 Å) | | 主引用文献 | Potential for Reduction of Streptogramin A Resistance Revealed by Structural Analysis of Acetyltransferase VatA.
Antimicrob.Agents Chemother., 58, 2014
|
|
1Y2K
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With 3,5-dimethyl-1-(3-nitro-phenyl)-1H-pyrazole-4-carboxylic acid ethyl ester | | 分子名称: | 1,2-ETHANEDIOL, 3,5-DIMETHYL-1-(3-NITROPHENYL)-1H-PYRAZOLE-4-CARBOXYLIC ACID ETHYL ESTER, MAGNESIUM ION, ... | | 著者 | Card, G.L, Blasdel, L, England, B.P, Zhang, C, Suzuki, Y, Gillette, S, Fong, D, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | 登録日 | 2004-11-22 | | 公開日 | 2005-03-01 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.36 Å) | | 主引用文献 | A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design
Nat.Biotechnol., 23, 2005
|
|
4A12
 
 | | Structure of the global transcription regulator FapR from Staphylococcus aureus in complex with DNA operator | | 分子名称: | FAPR PROMOTER, TRANSCRIPTION FACTOR FAPR | | 著者 | Albanesi, D, Guerin, M.E, Buschiazzo, A, de Mendoza, D, Alzari, P.M. | | 登録日 | 2011-09-13 | | 公開日 | 2012-09-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Structural Basis for Feed-Forward Transcriptional Regulation of Membrane Lipid Homeostasis in Staphylococcus Aureus.
Plos Pathog., 9, 2013
|
|
2WKW
 
 | | Alcaligenes esterase complexed with product analogue | | 分子名称: | CARBOXYLESTERASE, GLYCEROL, SULFATE ION, ... | | 著者 | Bourne, P.C, Isupov, M.N, Littlechild, J.A. | | 登録日 | 2009-06-18 | | 公開日 | 2009-06-30 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.03 Å) | | 主引用文献 | The Atomic-Resolution Structure of a Novel Bacterial Esterase.
Structure, 8, 2000
|
|
5QGO
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with OX-220 | | 分子名称: | (3aR,4R,6R,7R,8aR)-6-phenyloctahydro-1H-3a,7-epiminocyclohepta[c]pyrrol-4-ol, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
5QGZ
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000161 | | 分子名称: | ACETATE ION, DIMETHYL SULFOXIDE, N-(3-chlorophenyl)-2-(4-fluorophenyl)acetamide, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4GEK
 
 | | Crystal Structure of wild-type CmoA from E.coli | | 分子名称: | (2S)-4-[{[(2S,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-3,4-dihydroxytetrahydrofuran-2-yl]methyl}(carboxylatomethyl)sulfonio] -2-ammoniobutanoate, SULFATE ION, tRNA (cmo5U34)-methyltransferase | | 著者 | Kim, J, Toro, R, Bonanno, J.B, Bhosle, R, Sampathkumar, P, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | 登録日 | 2012-08-02 | | 公開日 | 2012-10-10 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure-guided discovery of the metabolite carboxy-SAM that modulates tRNA function
Nature, 498, 2013
|
|
5QHF
 
 | | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands) -- Crystal Structure of NUDT7 in complex with NUOOA000301a | | 分子名称: | 2-(4-methoxyphenyl)-N-{5-[2-oxo-2-(3-oxopiperazin-1-yl)ethoxy]pyridin-3-yl}acetamide, ACETATE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Krojer, T, Talon, R, Fairhead, M, Diaz Saez, L, Bradley, A.R, Aimon, A, Collins, P, Brandao-Neto, J, Douangamath, A, Ruda, G.F, Szommer, T, Srikannathasan, V, Elkins, J, Spencer, J, London, N, Nelson, A, Brennan, P.E, Huber, K, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | 登録日 | 2018-05-15 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | PanDDA analysis group deposition of models with modelled events (e.g. bound ligands)
To Be Published
|
|
4N7L
 
 | |
4RUR
 
 | | Yeast 20S proteasome in complex with the alkaloid indolo-phakellin (4) | | 分子名称: | (2E,3aR,14aS)-9-bromo-2-imino-1,2,3,5,6,14a-hexahydro-4H,8H-imidazo[4',5':5,6]pyrrolo[1',2':4,5]pyrazino[1,2-a]indol-8-one, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | 著者 | Beck, P, Lansdell, T.A, Hewlett, N.M, Tepe, J.J, Groll, M. | | 登録日 | 2014-11-21 | | 公開日 | 2014-12-10 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Indolo-Phakellins as beta 5-Specific Noncovalent Proteasome Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
