5RHF
 
 | | PanDDA analysis group deposition SARS-CoV-2 main protease fragment screen -- Crystal Structure of SARS-CoV-2 main protease in complex with PG-COV-34 (Mpro-x2754) | | 分子名称: | 1-acetyl-N-methyl-N-phenylpiperidine-4-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-05-16 | | 公開日 | 2020-06-10 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (1.76 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
2JG8
 
 | | Crystallographic structure of human C1q globular heads complexed to phosphatidyl-serine | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Complement C1q subcomponent subunit A, ... | | 著者 | Paidassi, H, Tacnet-Delorme, P, Garlatti, V, Darnault, C, Ghebrehiwet, B, Gaboriaud, C, Arlaud, G.J, Frachet, P. | | 登録日 | 2007-02-09 | | 公開日 | 2008-02-19 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | C1Q Binds Phosphatidylserine and Likely Acts as a Multiligand-Bridging Molecule in Apoptotic Cell Recognition.
J.Immunol., 180, 2008
|
|
6ADE
 
 | | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution | | 分子名称: | Glyceraldehyde-3-phosphate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Dilawari, R, Singh, P.K, Raje, M, Sharma, S, Singh, T.P. | | 登録日 | 2018-07-31 | | 公開日 | 2018-08-22 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (3.15 Å) | | 主引用文献 | Crystal structure of phosphorylated mutant of glyceraldehyde 3-phosphate dehydrogenase from human placenta at 3.15A resolution
To Be Published
|
|
2JGX
 
 | | Structure of CCP module 7 of complement factor H - The AMD Not at risk varient (402Y) | | 分子名称: | COMPLEMENT FACTOR H | | 著者 | Herbert, A.P, Deakin, J.A, Schmidt, C.Q, Blaum, B.S, Egan, C, Ferreira, V.P, Pangburn, M.K, Lyon, M, Uhrin, D, Barlow, P.N. | | 登録日 | 2007-02-16 | | 公開日 | 2007-03-20 | | 最終更新日 | 2018-05-02 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure shows that a glycosaminoglycan and protein recognition site in factor H is perturbed by age-related macular degeneration-linked single nucleotide polymorphism.
J. Biol. Chem., 282, 2007
|
|
4HQJ
 
 | | Crystal structure of Na+,K+-ATPase in the Na+-bound state | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, CHOLESTEROL, MAGNESIUM ION, ... | | 著者 | Nyblom, M, Reinhard, L, Gourdon, P, Nissen, P. | | 登録日 | 2012-10-25 | | 公開日 | 2013-10-02 | | 最終更新日 | 2014-09-10 | | 実験手法 | X-RAY DIFFRACTION (4.3 Å) | | 主引用文献 | Crystal structure of Na+, K(+)-ATPase in the Na(+)-bound state.
Science, 342, 2013
|
|
3QCY
 
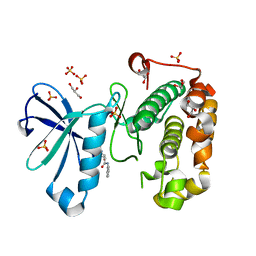 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 4-[2-Amino-6-(3-amino-1H-indazol-6-yl)-4-pyrimidinyl]-N-phenyl-2-morpholinecarboxamide | | 分子名称: | (2S)-4-[2-amino-6-(3-amino-2H-indazol-6-yl)pyrimidin-4-yl]-N-phenylmorpholine-2-carboxamide, 3-phosphoinositide-dependent protein kinase 1, GLYCEROL, ... | | 著者 | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | 登録日 | 2011-01-17 | | 公開日 | 2011-03-09 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
1E3Z
 
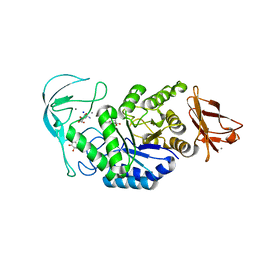 | | Acarbose complex of chimaeric amylase from B. amyloliquefaciens and B. licheniformis at 1.93A | | 分子名称: | 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose, 4,6-dideoxy-alpha-D-xylo-hexopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 6-AMINO-4-HYDROXYMETHYL-CYCLOHEX-4-ENE-1,2,3-TRIOL, ... | | 著者 | Brzozowski, A.M, Lawson, D.M, Turkenburg, J.P, Bisgaard-Frantzen, H, Svendsen, A, Borchert, T.V, Dauter, Z, Wilson, K.S, Davies, G.J. | | 登録日 | 2000-06-27 | | 公開日 | 2001-06-21 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.93 Å) | | 主引用文献 | Structural Analysis of a Chimeric Bacterial Alpha-Amylase. High Resolution Analysis of Native and Ligand Complexes
Biochemistry, 39, 2000
|
|
4NPY
 
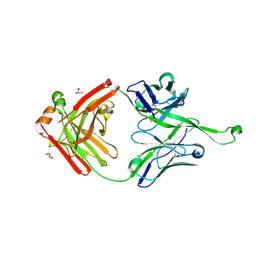 | | Crystal structure of germline Fab PGT121, a putative precursor of the broadly reactive and potent HIV-1 neutralizing antibody | | 分子名称: | GLYCEROL, germline PGT121 heavy chain, germline PGT121 light chain | | 著者 | Julien, J.-P, Diwanji, D.C, Wilson, I.A. | | 登録日 | 2013-11-22 | | 公開日 | 2013-12-11 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.796 Å) | | 主引用文献 | The Effects of Somatic Hypermutation on Neutralization and Binding in the PGT121 Family of Broadly Neutralizing HIV Antibodies.
Plos Pathog., 9, 2013
|
|
5ZZV
 
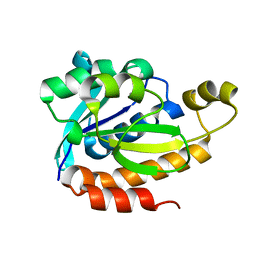 | | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution | | 分子名称: | 1,2-ETHANEDIOL, Peptidyl-tRNA hydrolase | | 著者 | Bairagya, H.R, Sharma, P, Iqbal, N, Sharma, S, Singh, T.P. | | 登録日 | 2018-06-04 | | 公開日 | 2018-06-20 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Crystal structure of PEG-1500 crystallized Peptidyl-tRNA Hydrolase from Acinetobacter baumannii at 1.5 A resolution
To Be Published
|
|
7Z28
 
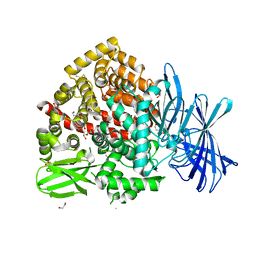 | | High-resolution crystal structure of ERAP1 with bound bestatin analogue inhibitor | | 分子名称: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Giastas, P, Papakyriakou, A, Stratikos, E, Vourloumis, D. | | 登録日 | 2022-02-26 | | 公開日 | 2022-07-27 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Discovery of Selective Nanomolar Inhibitors for Insulin-Regulated Aminopeptidase Based on alpha-Hydroxy-beta-amino Acid Derivatives of Bestatin.
J.Med.Chem., 65, 2022
|
|
4BQR
 
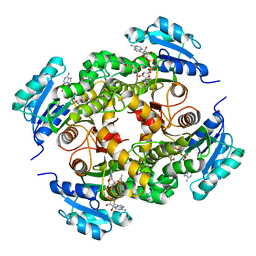 | | Mtb InhA complex with Methyl-thiazole compound 11 | | 分子名称: | (NZ)-2-[2,6-bis(fluoranyl)phenyl]-N-[5-[(1S)-1-(4-methyl-1,3-thiazol-2-yl)-1-oxidanyl-ethyl]-3H-1,3,4-thiadiazol-2-ylidene]ethanamide, ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | 著者 | Read, J.A, Gingell, H, Madhavapeddi, P, Shirude, P.S. | | 登録日 | 2013-05-31 | | 公開日 | 2013-12-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Methyl-Thiazoles: A Novel Mode of Inhibition with the Potential to Develop Novel Inhibitors Targeting Inha in Mycobacterium Tuberculosis.
J.Med.Chem., 56, 2013
|
|
5R84
 
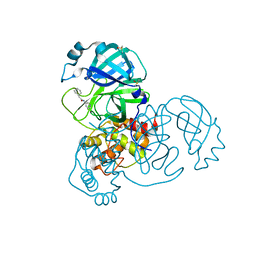 | | PanDDA analysis group deposition -- Crystal Structure of COVID-19 main protease in complex with Z31792168 | | 分子名称: | 2-cyclohexyl-~{N}-pyridin-3-yl-ethanamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Powell, A.J, Douangamath, A, Owen, C.D, Wild, C, Krojer, T, Lukacik, P, Strain-Damerell, C.M, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-03 | | 公開日 | 2020-03-11 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
3QCX
 
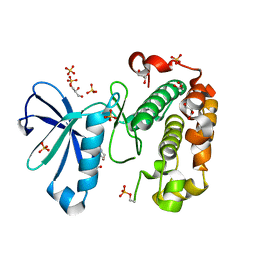 | | Phosphoinositide-Dependent Kinase-1 (PDK1) kinase domain with 6-{2-Amino-6-[(3R)-3-methyl-4-morpholinyl]-4-pyrimidinyl}-1H-indazol-3-amine | | 分子名称: | 3-phosphoinositide-dependent protein kinase 1, 6-{2-amino-6-[(3R)-3-methylmorpholin-4-yl]pyrimidin-4-yl}-2H-indazol-3-amine, GLYCEROL, ... | | 著者 | Medina, J.R, Becker, C.J, Blackledge, C.W, Duquenne, C, Feng, Y, Grant, S.W, Heerding, D, Li, W.H, Miller, W.H, Romeril, S.P, Scherzer, D, Shu, A, Bobko, M.A, Chadderton, A.R, Dumble, M, Gradiner, C.M, Gilbert, S, Liu, Q, Rabindran, S.K, Sudakin, V, Xiang, H, Brady, P.G, Campobasso, N, Ward, P, Axten, J.M. | | 登録日 | 2011-01-17 | | 公開日 | 2011-03-09 | | 最終更新日 | 2011-11-16 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure-Based Design of Potent and Selective 3-Phosphoinositide-Dependent Kinase-1 (PDK1) Inhibitors.
J.Med.Chem., 54, 2011
|
|
2JPA
 
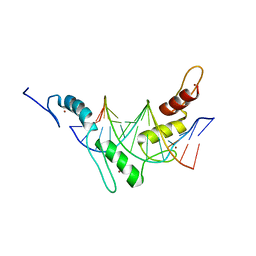 | | Structure of the Wilms Tumor Suppressor Protein Zinc Finger Domain Bound to DNA | | 分子名称: | DNA (5'-D(P*DCP*DAP*DGP*DAP*DCP*DGP*DCP*DCP*DCP*DCP*DCP*DGP*DCP*DG)-3'), DNA (5'-D(P*DCP*DGP*DCP*DGP*DGP*DGP*DGP*DGP*DCP*DGP*DTP*DCP*DTP*DG)-3'), Wilms tumor 1, ... | | 著者 | Stoll, R, Lee, B.M, Debler, E.W, Laity, J.H, Wilson, I.A, Dyson, H.J, Wright, P.E. | | 登録日 | 2007-05-01 | | 公開日 | 2007-10-30 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of the wilms tumor suppressor protein zinc finger domain bound to DNA
J.Mol.Biol., 372, 2007
|
|
3CFW
 
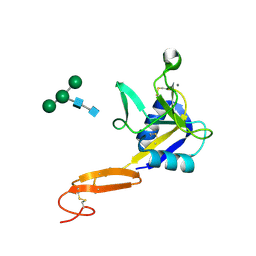 | | L-selectin lectin and EGF domains | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, L-selectin, ... | | 著者 | Mehta, P, Oganesyan, V, Terzyan, S, Mather, T, McEver, R.P. | | 登録日 | 2008-03-04 | | 公開日 | 2008-03-18 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Glycan Bound to the Selectin Low Affinity State Engages Glu-88 to Stabilize the High Affinity State under Force.
J.Biol.Chem., 292, 2017
|
|
1QZB
 
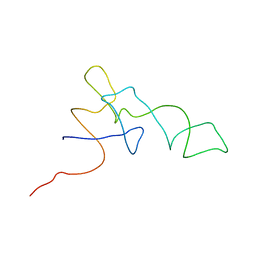 | | Coordinates of the A-site tRNA model fitted into the cryo-EM map of 70S ribosome in the pre-translocational state | | 分子名称: | Phe-tRNA | | 著者 | Valle, M, Zavialov, A, Li, W, Stagg, S.M, Sengupta, J, Nielsen, R.C, Nissen, P, Harvey, S.C, Ehrenberg, M, Frank, J. | | 登録日 | 2003-09-16 | | 公開日 | 2003-11-04 | | 最終更新日 | 2024-02-14 | | 実験手法 | ELECTRON MICROSCOPY (9 Å) | | 主引用文献 | Incorporation of Aminoacyl-tRNA into the Ribosome as seen by Cryo-electron Microscopy
Nat.Struct.Biol., 10, 2003
|
|
3LDH
 
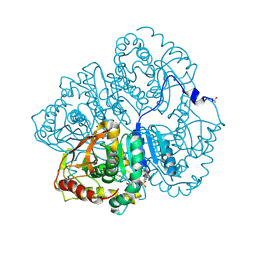 | | A comparison of the structures of apo dogfish m4 lactate dehydrogenase and its ternary complexes | | 分子名称: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID | | 著者 | White, J.L, Hackert, M.L, Buehner, M, Adams, M.J, Ford, G.C, Lentzjunior, P.J, Smiley, I.E, Steindel, S.J, Rossmann, M.G. | | 登録日 | 1974-06-06 | | 公開日 | 1977-04-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes.
J.Mol.Biol., 102, 1976
|
|
5RE8
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z2737076969 | | 分子名称: | 1-(3-fluorophenyl)-N-[(furan-2-yl)methyl]methanamine, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
5REN
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with PCM-0102425 | | 分子名称: | 1-[(3R)-3-(1,3-benzothiazol-2-yl)piperidin-1-yl]ethan-1-one, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2021-02-24 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
4NP4
 
 | | Clostridium difficile toxin B CROP domain in complex with FAB domains of neutralizing antibody bezlotoxumab | | 分子名称: | Toxin B, bezlotoxumab heavy chain, bezlotoxumab light chain | | 著者 | Orth, P, Xiao, L, Hernandez, L.D, Reichert, P, Sheth, P, Beaumont, M, Murgolo, N, Ermakov, G, DiNunzio, E, Racine, F, Karczewski, J, Secore, S, Ingram, R.N, Mayhood, T, Strickland, C, Therien, A.G. | | 登録日 | 2013-11-20 | | 公開日 | 2014-05-21 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.89 Å) | | 主引用文献 | Mechanism of Action and Epitopes of Clostridium difficile Toxin B-neutralizing Antibody Bezlotoxumab Revealed by X-ray Crystallography.
J.Biol.Chem., 289, 2014
|
|
4KA7
 
 | | Structure of Organellar OligoPeptidase (E572Q) in complex with an endogenous substrate | | 分子名称: | CHLORIDE ION, GLYCEROL, Oligopeptidase A, ... | | 著者 | Berntsson, R.P.-A, Kmiec, B, Teixeira, P.F, Svensson, L.M, Bakali, A, Glaser, E, Stenmark, P. | | 登録日 | 2013-04-22 | | 公開日 | 2013-09-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts.
Proc. Natl. Acad. Sci. U.S.A., 110, 2013
|
|
1QWM
 
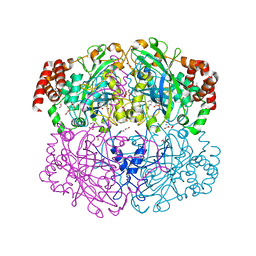 | | Structure of Helicobacter pylori catalase with formic acid bound | | 分子名称: | AZIDE ION, FORMIC ACID, KatA catalase, ... | | 著者 | Loewen, P.C, Carpena, X, Perez-Luque, R, Rovira, C, Haas, R, Odenbreit, S, Nicholls, P, Fita, I. | | 登録日 | 2003-09-02 | | 公開日 | 2004-03-30 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Structure of Helicobacter pylori Catalase, with and without Formic Acid Bound, at 1.6 A Resolution
Biochemistry, 43, 2004
|
|
6AM7
 
 | |
1B9K
 
 | |
5REG
 
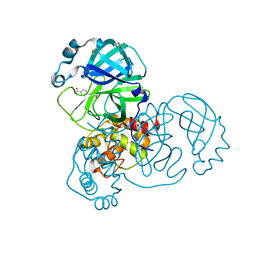 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 main protease in complex with Z1545313172 | | 分子名称: | (2~{S})-~{N}-(4-aminocarbonylphenyl)oxolane-2-carboxamide, 3C-like proteinase, DIMETHYL SULFOXIDE | | 著者 | Fearon, D, Owen, C.D, Douangamath, A, Lukacik, P, Powell, A.J, Strain-Damerell, C.M, Resnick, E, Krojer, T, Gehrtz, P, Wild, C, Aimon, A, Brandao-Neto, J, Carbery, A, Dunnett, L, Skyner, R, Snee, M, London, N, Walsh, M.A, von Delft, F. | | 登録日 | 2020-03-15 | | 公開日 | 2020-03-25 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Crystallographic and electrophilic fragment screening of the SARS-CoV-2 main protease.
Nat Commun, 11, 2020
|
|
