7AFZ
 
 | | L1 metallo-b-lactamase with compound EBL-1306 | | 分子名称: | 3-[3-chloranyl-4-(methylsulfonylmethyl)phenyl]-7-propan-2-yl-1~{H}-indole-2-carboxylic acid, Metallo-beta-lactamase L1, SULFATE ION, ... | | 著者 | Hinchliffe, P, Spencer, J, Brem, J. | | 登録日 | 2020-09-21 | | 公開日 | 2021-10-06 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Imitation of beta-lactam binding enables broad-spectrum metallo-beta-lactamase inhibitors.
Nat.Chem., 14, 2022
|
|
7ABU
 
 | | Structure of SARS-CoV-2 Main Protease bound to RS102895 | | 分子名称: | 1'-[2-[4-(trifluoromethyl)phenyl]ethyl]spiro[1~{H}-3,1-benzoxazine-4,4'-piperidine]-2-one, 3C-like proteinase, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Lane, T.J, Dunkel, I, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-08 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
6PFK
 
 | |
7ADW
 
 | | Structure of SARS-CoV-2 Main Protease bound to 2,4'-Dimethylpropiophenone. | | 分子名称: | 2-methyl-1-(4-methylphenyl)propan-1-one, 3C-like proteinase, CHLORIDE ION, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-09-16 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.63 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7A1U
 
 | | Structure of SARS-CoV-2 Main Protease bound to Fusidic Acid. | | 分子名称: | 3C-like proteinase, DIMETHYL SULFOXIDE, FUSIDIC ACID, ... | | 著者 | Guenther, S, Reinke, P, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Paulraj, L.X, Ullah, N, Andaleeb, H, Werner, N, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Doyle, J.J, Giseler, H, Melo, D, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2020-08-14 | | 公開日 | 2020-12-02 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7QGQ
 
 | |
8GKH
 
 | | Structure of the Spizellomyces punctatus Fanzor (SpuFz) in complex with omega RNA and target DNA | | 分子名称: | DNA (35-MER), DNA (5'-D(P*CP*GP*GP*TP*AP*CP*CP*CP*GP*GP*GP*CP*AP*TP*A)-3'), MAGNESIUM ION, ... | | 著者 | Xu, P, Saito, M, Zhang, F. | | 登録日 | 2023-03-19 | | 公開日 | 2023-07-05 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | Fanzor is a eukaryotic programmable RNA-guided endonuclease.
Nature, 620, 2023
|
|
7QIX
 
 | | Specific features and methylation sites of a plant ribosome. 40S body ribosomal subunit. | | 分子名称: | 18S rRNA body, 30S ribosomal protein S15, chloroplastic, ... | | 著者 | Cottilli, P, Itoh, Y, Amunts, A. | | 登録日 | 2021-12-16 | | 公開日 | 2022-06-15 | | 最終更新日 | 2024-04-24 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | Cryo-EM structure and rRNA modification sites of a plant ribosome.
Plant Commun., 3, 2022
|
|
7Q7Q
 
 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | 著者 | Baath, P, Banacore, A, Neutze, R. | | 登録日 | 2021-11-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7Q7P
 
 | | LIPIDIC CUBIC PHASE SERIAL FEMTOSECOND CRYSTALLOGRAPHY STRUCTURE OF A PHOTOSYNTHETIC REACTION CENTRE | | 分子名称: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | 著者 | Baath, P, Banacore, A, Neutze, R. | | 登録日 | 2021-11-09 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Lipidic cubic phase serial femtosecond crystallography structure of a photosynthetic reaction centre.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7AAP
 
 | | Nsp7-Nsp8-Nsp12 SARS-CoV2 RNA-dependent RNA polymerase in complex with template:primer dsRNA and favipiravir-RTP | | 分子名称: | MAGNESIUM ION, Non-structural protein 12, Non-structural protein 7, ... | | 著者 | Naydenova, K, Muir, K.W, Wu, L.F, Zhang, Z, Coscia, F, Peet, M, Castro-Hartman, P, Qian, P, Sader, K, Dent, K, Kimanius, D, Sutherland, J.D, Lowe, J, Barford, D, Russo, C.J. | | 登録日 | 2020-09-04 | | 公開日 | 2020-09-23 | | 最終更新日 | 2024-07-10 | | 実験手法 | ELECTRON MICROSCOPY (2.5 Å) | | 主引用文献 | Structure of the SARS-CoV-2 RNA-dependent RNA polymerase in the presence of favipiravir-RTP.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7ABP
 
 | |
8BHD
 
 | | N-terminal domain of Plasmodium berghei glutamyl-tRNA synthetase (Tbxo4 derivative crystal structure) | | 分子名称: | GLYCEROL, Glutamate--tRNA ligase, SULFATE ION, ... | | 著者 | Benas, P, Jaramillo Ponce, J.R, Legrand, P, Frugier, M, Sauter, C. | | 登録日 | 2022-10-31 | | 公開日 | 2023-01-25 | | 最終更新日 | 2024-06-19 | | 実験手法 | X-RAY DIFFRACTION (3.17 Å) | | 主引用文献 | Solution X-ray scattering highlights discrepancies in Plasmodium multi-aminoacyl-tRNA synthetase complexes.
Protein Sci., 32, 2023
|
|
6ZH7
 
 | | Crystal structure of fatty acid photodecarboxylase in the dark state determined by serial femtosecond crystallography at room temperature | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, Fatty acid photodecarboxylase, chloroplastic, ... | | 著者 | Hadjidemetriou, K, Coquelle, N, Weik, M, Schlichting, I, Barends, T.R.M, Colletier, J.P. | | 登録日 | 2020-06-21 | | 公開日 | 2021-04-14 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Mechanism and dynamics of fatty acid photodecarboxylase.
Science, 372, 2021
|
|
8F43
 
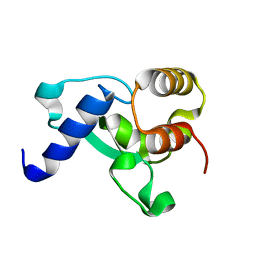 | | HNH Nuclease Domain from G. stearothermophilus Cas9, K597A mutant | | 分子名称: | CRISPR-associated endonuclease Cas9 | | 著者 | D'Ordine, A.M, Belato, H.B, Lisi, G.P, Jogl, G. | | 登録日 | 2022-11-10 | | 公開日 | 2022-12-21 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (1.37 Å) | | 主引用文献 | Disruption of electrostatic contacts in the HNH nuclease from a thermophilic Cas9 rewires allosteric motions and enhances high-temperature DNA cleavage.
J.Chem.Phys., 157, 2022
|
|
6OCQ
 
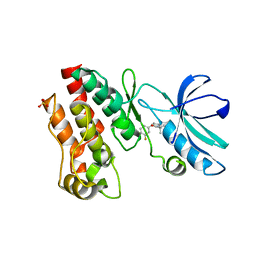 | | Crystal structure of RIP1 kinase in complex with a pyrrolidine | | 分子名称: | 1-[(2S)-2-(3-fluorophenyl)pyrrolidin-1-yl]-2,2-dimethylpropan-1-one, Receptor-interacting serine/threonine-protein kinase 1, SULFATE ION | | 著者 | Thorpe, J.H, Harris, P.A. | | 登録日 | 2019-03-25 | | 公開日 | 2019-05-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.793 Å) | | 主引用文献 | Discovery and Lead-Optimization of 4,5-Dihydropyrazoles as Mono-Kinase Selective, Orally Bioavailable and Efficacious Inhibitors of Receptor Interacting Protein 1 (RIP1) Kinase.
J.Med.Chem., 62, 2019
|
|
7YI4
 
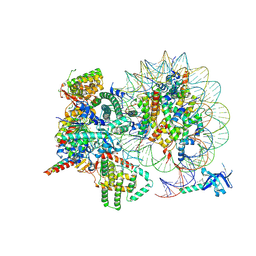 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in close state | | 分子名称: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
7YI5
 
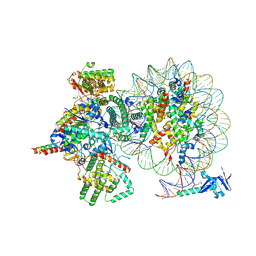 | | Cryo-EM structure of Rpd3S complex bound to H3K36me3 nucleosome in loose state | | 分子名称: | Chromatin modification-related protein EAF3, Histone H2A, Histone H2B 1.1, ... | | 著者 | Li, H.T, Yan, C.Y, Guan, H.P, Wang, P. | | 登録日 | 2022-07-14 | | 公開日 | 2023-06-14 | | 最終更新日 | 2023-08-30 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Diverse modes of H3K36me3-guided nucleosomal deacetylation by Rpd3S.
Nature, 620, 2023
|
|
8EQI
 
 | |
8EL5
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in an alternating tight to loose interface filament | | 分子名称: | DiCys-(15,16)-Dihydrobiliverdin, GLYCEROL, PHYCOERYTHROBILIN, ... | | 著者 | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | 登録日 | 2022-09-23 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.67 Å) | | 主引用文献 | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL4
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in a tight interface filament | | 分子名称: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | 著者 | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | 登録日 | 2022-09-23 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL3
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 in a loose interface filament | | 分子名称: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | 著者 | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | 登録日 | 2022-09-23 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.57 Å) | | 主引用文献 | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
8EL6
 
 | | Light harvesting phycobiliprotein HaPE555 from the cryptophyte Hemiselmis andersenii CCMP644 with an altered helix hA/hY conformation | | 分子名称: | DiCys-(15,16)-Dihydrobiliverdin, PHYCOERYTHROBILIN, Phycoerythrin alpha-1 subunit, ... | | 著者 | Rathbone, H.W, Michie, K.A, Laos, A.L, Curmi, P.M.G. | | 登録日 | 2022-09-23 | | 公開日 | 2023-10-25 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Molecular dissection of the soluble photosynthetic antenna from the cryptophyte alga Hemiselmis andersenii.
Commun Biol, 6, 2023
|
|
7NEV
 
 | | Structure of the hemiacetal complex between the SARS-CoV-2 Main Protease and Leupeptin | | 分子名称: | 3C-like proteinase, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | 著者 | Guenther, S, Reinke, P.Y.A, Oberthuer, D, Yefanov, O, Gelisio, L, Ginn, H.M, Lieske, J, Domaracky, M, Brehm, W, Rahmani Mashhour, A, White, T.A, Knoska, J, Pena Esperanza, G, Koua, F, Tolstikova, A, Groessler, M, Fischer, P, Hennicke, V, Fleckenstein, H, Trost, F, Galchenkova, M, Gevorkov, Y, Li, C, Awel, S, Xavier, P.L, Ullah, N, Andaleeb, H, Falke, S, Alves Franca, B, Schwinzer, M, Brognaro, H, Werner, N, Perbandt, M, Tidow, H, Seychell, B, Beck, T, Meier, S, Zaitsev-Doyle, J.J, Rogers, C, Gieseler, H, Melo, D, Monteiro, D.C.F, Dunkel, I, Lane, T.J, Peck, A, Saouane, S, Hakanpaeae, J, Meyer, J, Noei, H, Gribbon, P, Ellinger, B, Kuzikov, M, Wolf, M, Zhang, L, Ehrt, C, Pletzer-Zelgert, J, Wollenhaupt, J, Feiler, C, Weiss, M, Schluenzen, F, Schulz, E.C, Mehrabi, P, Norton-Baker, B, Schmidt, C, Lorenzen, K, Schubert, R, Sun, X, Han, H, Chari, A, Fernandez Garcia, Y, Turk, D, Hilgenfeld, R, Rarey, M, Zaliani, A, Chapman, H.N, Pearson, A, Betzel, C, Meents, A. | | 登録日 | 2021-02-05 | | 公開日 | 2021-03-03 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | X-ray screening identifies active site and allosteric inhibitors of SARS-CoV-2 main protease.
Science, 372, 2021
|
|
7B3D
 
 | | Structure of elongating SARS-CoV-2 RNA-dependent RNA polymerase with AMP at position -4 (structure 3) | | 分子名称: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase nsp12, ... | | 著者 | Kokic, G, Hillen, H.S, Tegunov, D, Dienemann, C, Seitz, F, Schmitzova, J, Farnung, L, Siewert, A, Hoebartner, C, Cramer, P. | | 登録日 | 2020-11-30 | | 公開日 | 2020-12-23 | | 最終更新日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.8 Å) | | 主引用文献 | Mechanism of SARS-CoV-2 polymerase stalling by remdesivir.
Nat Commun, 12, 2021
|
|
