4WX2
 
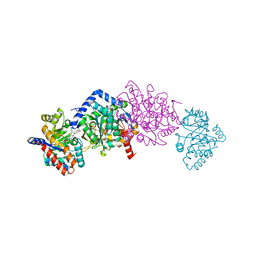 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium in complex with two F6F molecules in the alpha-site and one F6F molecule in the beta-site | | 分子名称: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Hilario, E, Caulkins, B.G, Young, R.P, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | 登録日 | 2014-11-13 | | 公開日 | 2015-11-18 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Visualizing the tunnel in tryptophan synthase with crystallography: Insights into a selective filter for accommodating indole and rejecting water.
Biochim.Biophys.Acta, 1864, 2016
|
|
1JQ5
 
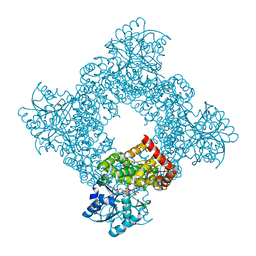 | | Bacillus Stearothermophilus Glycerol dehydrogenase complex with NAD+ | | 分子名称: | Glycerol dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ZINC ION | | 著者 | Ruzheinikov, S.N, Burke, J, Sedelnikova, S, Baker, P.J, Taylor, R, Bullough, P.A, Muir, N.M, Gore, M.G, Rice, D.W. | | 登録日 | 2001-08-03 | | 公開日 | 2001-10-01 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Glycerol dehydrogenase. structure, specificity, and mechanism of a family III polyol dehydrogenase.
Structure, 9, 2001
|
|
1B3C
 
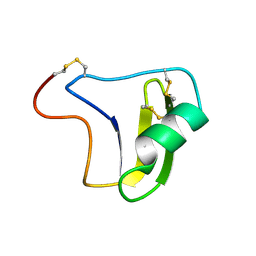 | | SOLUTION STRUCTURE OF A BETA-NEUROTOXIN FROM THE NEW WORLD SCORPION CENTRUROIDES SCULPTURATUS EWING | | 分子名称: | PROTEIN (NEUROTOXIN CSE-I) | | 著者 | Jablonsky, M.J, Jackson, P.L, Trent, J.O, Watt, D.D, Krishna, N.R. | | 登録日 | 1998-12-08 | | 公開日 | 1998-12-16 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a beta-neurotoxin from the New World scorpion Centruroides sculpturatus Ewing.
Biochem.Biophys.Res.Commun., 254, 1999
|
|
1BG5
 
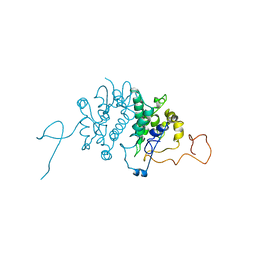 | | CRYSTAL STRUCTURE OF THE ANKYRIN BINDING DOMAIN OF ALPHA-NA,K-ATPASE AS A FUSION PROTEIN WITH GLUTATHIONE S-TRANSFERASE | | 分子名称: | FUSION PROTEIN OF ALPHA-NA,K-ATPASE WITH GLUTATHIONE S-TRANSFERASE | | 著者 | Zhang, Z, Devarajan, P, Morrow, J.S. | | 登録日 | 1998-06-05 | | 公開日 | 1999-01-13 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the ankyrin-binding domain of alpha-Na,K-ATPase.
J.Biol.Chem., 273, 1998
|
|
3S3W
 
 | | Structure of chicken acid-sensing ion channel 1 at 2.6 a resolution and ph 7.5 | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Amiloride-sensitive cation channel 2, ... | | 著者 | Dawson, R.J.P, Benz, J, Stohler, P, Tetaz, T, Joseph, C, Huber, S, Schmid, G, Huegin, D, Pflimlin, P, Trube, G, Rudolph, M.G, Hennig, M, Ruf, A. | | 登録日 | 2011-05-18 | | 公開日 | 2012-05-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of the Acid-sensing ion channel 1 in complex with the gating modifier Psalmotoxin 1.
Nat Commun, 3, 2012
|
|
1MWQ
 
 | | Structure of HI0828, a Hypothetical Protein from Haemophilus influenzae with a Putative Active-Site Phosphohistidine | | 分子名称: | CACODYLATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Willis, M.A, Krajewski, W, Chalamasetty, V.R, Reddy, P, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | 登録日 | 2002-09-30 | | 公開日 | 2003-11-25 | | 最終更新日 | 2019-07-24 | | 実験手法 | X-RAY DIFFRACTION (0.99 Å) | | 主引用文献 | Structure of YciI from Haemophilus influenzae (HI0828) reveals a ferredoxin-like alpha/beta-fold with a histidine/aspartate centered catalytic site
Proteins, 59, 2005
|
|
2RER
 
 | |
8IFF
 
 | | Cryo-EM structure of Arabidopsis phytochrome A. | | 分子名称: | 3-[5-[[(3~{R},4~{R})-3-ethyl-4-methyl-5-oxidanylidene-3,4-dihydropyrrol-2-yl]methyl]-2-[[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1~{H}-pyrrol-2-yl]methyl]-4-methyl-1~{H}-pyrrol-3-yl]propanoic acid, Phytochrome A | | 著者 | Ma, L, Zhou, C, Wang, J, Guan, Z, Yin, P. | | 登録日 | 2023-02-17 | | 公開日 | 2023-08-02 | | 最終更新日 | 2023-10-11 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Plant phytochrome A in the Pr state assembles as an asymmetric dimer.
Cell Res., 33, 2023
|
|
1BIK
 
 | | X-RAY STRUCTURE OF BIKUNIN FROM THE HUMAN INTER-ALPHA-INHIBITOR COMPLEX | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BIKUNIN, SULFATE ION | | 著者 | Xu, Y, Carr, P.D, Guss, J.M, Ollis, D.L. | | 登録日 | 1997-11-26 | | 公開日 | 1999-03-16 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | The crystal structure of bikunin from the inter-alpha-inhibitor complex: a serine protease inhibitor with two Kunitz domains.
J.Mol.Biol., 276, 1998
|
|
3VNM
 
 | | Crystal structures of D-Psicose 3-epimerase with D-sorbose from Clostridium cellulolyticum H10 | | 分子名称: | D-sorbose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-17 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.12 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
1K0M
 
 | | Crystal structure of a soluble monomeric form of CLIC1 at 1.4 angstroms | | 分子名称: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | 著者 | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | 登録日 | 2001-09-19 | | 公開日 | 2001-12-12 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
3F5C
 
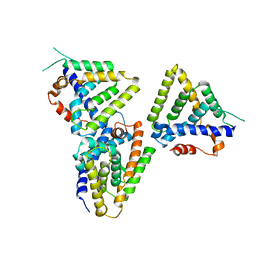 | | Structure of Dax-1:LRH-1 complex | | 分子名称: | Nuclear receptor subfamily 0 group B member 1, Nuclear receptor subfamily 5 group A member 2 | | 著者 | Fletterick, R.J, Sablin, E.P. | | 登録日 | 2008-11-03 | | 公開日 | 2008-12-23 | | 最終更新日 | 2023-09-06 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
6YPH
 
 | |
8HVI
 
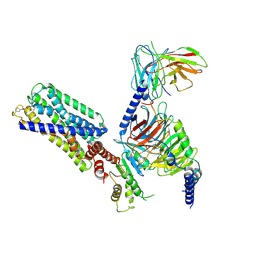 | | Activation mechanism of GPR132 by compound NOX-6-7 | | 分子名称: | 3-methyl-5-[(4-oxidanylidene-4-phenyl-butanoyl)amino]-1-benzofuran-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | 著者 | Wang, J.L, Ding, J.H, Sun, J.P, Yu, X. | | 登録日 | 2022-12-26 | | 公開日 | 2023-10-11 | | 最終更新日 | 2023-11-01 | | 実験手法 | ELECTRON MICROSCOPY (3.04 Å) | | 主引用文献 | Functional screening and rational design of compounds targeting GPR132 to treat diabetes.
Nat Metab, 5, 2023
|
|
7PCV
 
 | |
1B9U
 
 | |
7SZZ
 
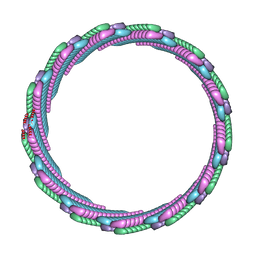 | | Structure of the smaller diameter PSMalpha3 nanotubes | | 分子名称: | Phenol-soluble modulin PSM-alpha-3 | | 著者 | Beltran, L.C, Kreutzberger, M.A, Wang, S, Egelman, E.H, Conticello, V.P. | | 登録日 | 2021-11-29 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (3.9 Å) | | 主引用文献 | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1EZT
 
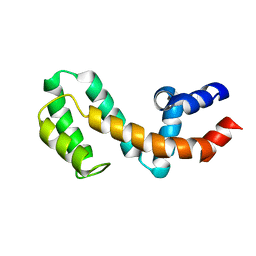 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF FREE RGS4 BY NMR | | 分子名称: | REGULATOR OF G-PROTEIN SIGNALING 4 | | 著者 | Moy, F.J, Chanda, P.K, Cockett, M.I, Edris, W, Jones, P.G, Mason, K, Semus, S, Powers, R. | | 登録日 | 2000-05-11 | | 公開日 | 2001-01-15 | | 最終更新日 | 2024-05-22 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of free RGS4 reveals an induced conformational change upon binding Galpha.
Biochemistry, 39, 2000
|
|
5YNJ
 
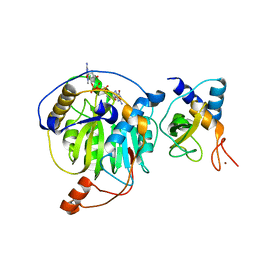 | | Crystal structure of MERS-CoV nsp16/nsp10 complex bound to m7GpppG | | 分子名称: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, ZINC ION, nsp10 protein, ... | | 著者 | Wei, S.M, Yang, L, Ke, Z.H, Chen, Y, Guo, D.Y, Fan, C.P. | | 登録日 | 2017-10-24 | | 公開日 | 2018-12-05 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.043 Å) | | 主引用文献 | Structural insights into the molecular mechanism of MERS Coronavirus RNA ribose 2'-O-methylation by nsp16/nsp10 protein complex
To Be Published
|
|
3VNL
 
 | | Crystal structures of D-Psicose 3-epimerase with D-tagatose from Clostridium cellulolyticum H10 | | 分子名称: | D-tagatose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3VNK
 
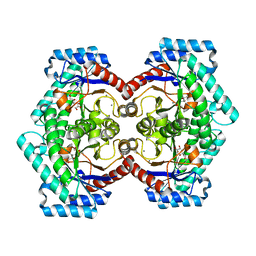 | | Crystal structures of D-Psicose 3-epimerase with D-fructose from Clostridium cellulolyticum H10 | | 分子名称: | D-fructose, MANGANESE (II) ION, Xylose isomerase domain protein TIM barrel | | 著者 | Chan, H.C, Zhu, Y, Hu, Y, Ko, T.P, Huang, C.H, Ren, F, Chen, C.C, Guo, R.T, Sun, Y. | | 登録日 | 2012-01-16 | | 公開日 | 2012-08-01 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.02 Å) | | 主引用文献 | Crystal structures of D-psicose 3-epimerase from Clostridium cellulolyticum H10 and its complex with ketohexose sugars.
Protein Cell, 3, 2012
|
|
3O4Q
 
 | |
6Y10
 
 | | X-ray structure of Lactobacillus brevis alcohol dehydrogenase mutant Q126H | | 分子名称: | 1,2-ETHANEDIOL, MAGNESIUM ION, R-specific alcohol dehydrogenase | | 著者 | Hermann, J, Bischoff, D, Janowski, R, Niessing, D, Grob, P, Hekmat, D, Weuster-Botz, D. | | 登録日 | 2020-02-10 | | 公開日 | 2020-02-19 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.22 Å) | | 主引用文献 | Controlling Protein Crystallization by Free Energy Guided Design of Interactions at Crystal Contacts
Crystals, 11, 2021
|
|
7T0X
 
 | | Structure of the larger diameter PSMalpha3 nanotube | | 分子名称: | Phenol-soluble modulin PSM-alpha-3 | | 著者 | Kreutzberger, M.A, Wang, S, Beltran, L.C, Egelman, E.H, Conticello, V.P. | | 登録日 | 2021-11-30 | | 公開日 | 2022-05-18 | | 最終更新日 | 2024-02-28 | | 実験手法 | ELECTRON MICROSCOPY (4.4 Å) | | 主引用文献 | Phenol-soluble modulins PSM alpha 3 and PSM beta 2 form nanotubes that are cross-alpha amyloids.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
5KKK
 
 | |
