5SYL
 
 | | B. pseudomallei KatG with KCN bound | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, CYANIDE ION, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-11 | | Release date: | 2016-09-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of B. pseudomallei KatG with cyanide bound
To be published
|
|
6KA6
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 1 | | Descriptor: | (3~{S})-3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)-3,4-dihydroisochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-21 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.891 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
6NVC
 
 | | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 6 | | Descriptor: | ATP-dependent dethiobiotin synthetase BioD, SULFATE ION, {(1S,2R)-2-[4-(carboxymethyl)benzene-1-carbonyl]cyclopentyl}acetic acid | | Authors: | Thompson, A.P, Polyak, S.W, Wegener, K.L, Bruning, J.B. | | Deposit date: | 2019-02-04 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of Mycobacterium tuberculosis dethiobiotin synthetase in complex with fragment analogue 6
To Be Published
|
|
5SZE
 
 | | Crystal structure of Aquifex aeolicus Hfq-RNA complex at 1.5A | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, DI(HYDROXYETHYL)ETHER, RNA (5'-R(P*UP*UP*U)-3'), ... | | Authors: | Stanek, K, Patterson, J, Randolph, P.S, Mura, C. | | Deposit date: | 2016-08-13 | | Release date: | 2017-04-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure and RNA-binding properties of an Hfq homolog from the deep-branching Aquificae: conservation of the lateral RNA-binding mode.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
5SX2
 
 | | Crystal structure of the D141E mutant of B. pseudomallei KatG at pH 8.0. | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Catalase-peroxidase, ... | | Authors: | Loewen, P.C. | | Deposit date: | 2016-08-09 | | Release date: | 2016-09-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Two alternative substrate paths for compound I formation and reduction in catalase-peroxidase KatG from Burkholderia pseudomallei.
Proteins, 66, 2007
|
|
6KCN
 
 | | Crystal structure of plasmodium lysyl-tRNA synthetase in complex with a cladosporin derivative 4 | | Descriptor: | 3-[[(1~{S},3~{S})-3-methylcyclohexyl]methyl]-6,8-bis(oxidanyl)isochromen-1-one, GLYCEROL, LYSINE, ... | | Authors: | Zhou, J, Fang, P. | | Deposit date: | 2019-06-28 | | Release date: | 2020-04-08 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Atomic Resolution Analyses of Isocoumarin Derivatives for Inhibition of Lysyl-tRNA Synthetase.
Acs Chem.Biol., 15, 2020
|
|
2F43
 
 | | Rat liver F1-ATPase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP synthase alpha chain, ... | | Authors: | Chen, C, Saxena, A.K, Simcoke, W.N, Garboczi, D.N, Pedersen, P.L, Ko, Y.H. | | Deposit date: | 2005-11-22 | | Release date: | 2006-03-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Mitochondrial ATP synthase: Crystal structure of the catalytic F1 unit in a vanadate-induced transition-like state and implications for mechanism.
J.Biol.Chem., 281, 2006
|
|
5HJR
 
 | | Murine endoplasmic reticulum alpha-glucosidase II with bound covalent intermediate | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoro-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Caputo, A.T, Roversi, P, Alonzi, D.S, Kiappes, J.L, Zitzmann, N. | | Deposit date: | 2016-01-13 | | Release date: | 2016-07-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structures of mammalian ER alpha-glucosidase II capture the binding modes of broad-spectrum iminosugar antivirals.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
3LU6
 
 | | Human serum albumin in complex with compound 1 | | Descriptor: | Serum albumin, [(1R,2R)-2-{[(5-fluoro-1H-indol-2-yl)carbonyl]amino}-2,3-dihydro-1H-inden-1-yl]acetic acid | | Authors: | Buttar, D, Colclough, N, Gerhardt, S, MacFaul, P.A, Phillips, S.D, Plowright, A, Whittamore, P, Tam, K, Maskos, K, Steinbacher, S, Steuber, H. | | Deposit date: | 2010-02-17 | | Release date: | 2010-10-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A combined spectroscopic and crystallographic approach to probing drug-human serum albumin interactions
Bioorg.Med.Chem., 18, 2010
|
|
4BRG
 
 | | Legionella pneumophila NTPDase1 crystal form II (closed) in complex with MG GMPPNP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ECTONUCLEOSIDE TRIPHOSPHATE DIPHOSPHOHYDROLASE I, ... | | Authors: | Zebisch, M, Schaefer, P, Lauble, P, Straeter, N. | | Deposit date: | 2013-06-04 | | Release date: | 2013-07-17 | | Last modified: | 2013-12-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystallographic Snapshots Along the Reaction Pathway of Nucleoside Triphosphate Diphosphohydrolases
Structure, 21, 2013
|
|
5SYY
 
 | |
1AJ7
 
 | | IMMUNOGLOBULIN 48G7 GERMLINE FAB ANTIBODY COMPLEXED WITH HAPTEN 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID. AFFINITY MATURATION OF AN ESTEROLYTIC ANTIBODY | | Descriptor: | 5-(PARA-NITROPHENYL PHOSPHONATE)-PENTANOIC ACID, IMMUNOGLOBULIN 48G7 FAB (HEAVY CHAIN), IMMUNOGLOBULIN 48G7 FAB (LIGHT CHAIN) | | Authors: | Wedemayer, G.J, Wang, L.H, Patten, P.A, Schultz, P.G, Stevens, R.C. | | Deposit date: | 1997-05-15 | | Release date: | 1997-11-12 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the evolution of an antibody combining site.
Science, 276, 1997
|
|
5HIX
 
 | | Cocrystal structure of an anti-parallel DNA G-quadruplex and a tetra-Quinoline Foldamer | | Descriptor: | 4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-({[4-(3-aminopropoxy)-8-nitroquinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinolin-2-yl]carbonyl}amino)quinoline-2-carboxylic acid, Dimeric G-quadruplex, POTASSIUM ION | | Authors: | Mandal, P.K, Baptiste, B, Langlois d'Estaintot, B, Kauffmann, B, Huc, I. | | Deposit date: | 2016-01-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Multivalent Interactions between an Aromatic Helical Foldamer and a DNA G-Quadruplex in the Solid State.
Chembiochem, 17, 2016
|
|
1VEA
 
 | | Crystal Structure of HutP, an RNA binding antitermination protein | | Descriptor: | Hut operon positive regulatory protein, N-(2-NAPHTHYL)HISTIDINAMIDE | | Authors: | Kumarevel, T.S, Fujimoto, Z, Karthe, P, Oda, M, Mizuno, H, Kumar, P.K.R. | | Deposit date: | 2004-03-29 | | Release date: | 2004-07-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of Activated HutP; An RNA Binding Protein that Regulates Transcription of the hut Operon in Bacillus subtilis
Structure, 12, 2004
|
|
6O28
 
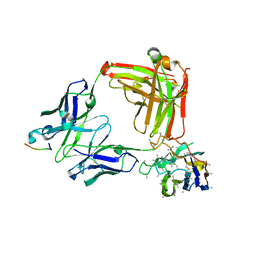 | | Crystal structure of 4493 Fab in complex with circumsporozoite protein KQPA and anti-kappa VHH domain | | Descriptor: | 1,2-ETHANEDIOL, 4493 Fab heavy chain, 4493 Kappa light chain, ... | | Authors: | Scally, S.W, Bosch, A, Prieto, K, Murugan, R, Wardemann, H, Julien, J.P. | | Deposit date: | 2019-02-22 | | Release date: | 2020-07-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Evolution of protective human antibodies against Plasmodium falciparum circumsporozoite protein repeat motifs.
Nat. Med., 26, 2020
|
|
3CG1
 
 | |
5T1L
 
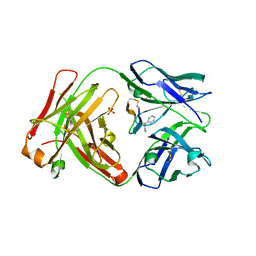 | | Cetuximab Fab in complex with CQA(Ph)2DLSTRRLKC peptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CETUXIMAB FAB HEAVY CHAIN, CETUXIMAB FAB LIGHT CHAIN, ... | | Authors: | Bzymek, K.P, Williams, J.C. | | Deposit date: | 2016-08-19 | | Release date: | 2016-10-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Natural and non-natural amino-acid side-chain substitutions: affinity and diffraction studies of meditope-Fab complexes.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
7XTV
 
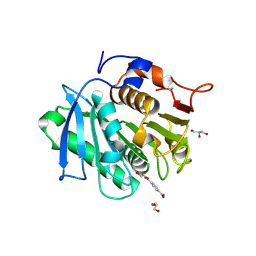 | | The structure of MHET-bound TfCut S130A | | Descriptor: | 4-(2-hydroxyethyloxycarbonyl)benzoic acid, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
7XTU
 
 | | The structure of TfCut S130A | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yang, Y, Jiang, P.C, Huang, J.-W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-05-18 | | Release date: | 2023-03-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Complete bio-degradation of poly(butylene adipate-co-terephthalate) via engineered cutinases.
Nat Commun, 14, 2023
|
|
2GMV
 
 | | PEPCK complex with a GTP-competitive inhibitor | | Descriptor: | MANGANESE (II) ION, N-(4-{[3-BUTYL-1-(2-FLUOROBENZYL)-2,6-DIOXO-2,3,6,7-TETRAHYDRO-1H-PURIN-8-YL]METHYL}PHENYL)-1-METHYL-1H-IMIDAZOLE-4-SULFONAMIDE, PHOSPHOENOLPYRUVATE, ... | | Authors: | Dunten, P. | | Deposit date: | 2006-04-07 | | Release date: | 2007-05-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | C-8 Modifications of 3-alkyl-1,8-dibenzylxanthines as inhibitors of human cytosolic phosphoenolpyruvate carboxykinase.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5T6C
 
 | |
4CBT
 
 | | Design, synthesis, and biological evaluation of potent and selective Class IIa HDAC inhibitors as a potential therapy for Huntington's disease | | Descriptor: | (1R,2R,3R)-2-[4-(5-fluoranylpyrimidin-2-yl)phenyl]-N-oxidanyl-3-phenyl-cyclopropane-1-carboxamide, HISTONE DEACETYLASE 4, ZINC ION | | Authors: | Burli, R.W, Luckhurst, C.A, Aziz, O, Matthews, K.L, Yates, D, Lyons, K.A, Beconi, M, McAllister, G, Breccia, P, Stott, A.J, Penrose, S.D, Wall, M, Lamers, M, Leonard, P, Mueller, I, Richardson, C.M, Jarvis, R, Stones, L, Hughes, S, Wishart, G, Haughan, A.F, O'Connell, C, Mead, T, McNeil, H, Vann, J, Mangette, J, Maillard, M, Beaumont, V, Munoz-Sanjuan, I, Dominguez, C. | | Deposit date: | 2013-10-16 | | Release date: | 2013-12-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Design, synthesis, and biological evaluation of potent and selective class IIa histone deacetylase (HDAC) inhibitors as a potential therapy for Huntington's disease.
J. Med. Chem., 56, 2013
|
|
4TOY
 
 | |
4JV7
 
 | | Co-crystal structure of MDM2 with inhibitor (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one | | Descriptor: | (2S,5R,6S)-2-benzyl-5,6-bis(4-bromophenyl)-4-methylmorpholin-3-one, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-25 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
4JVR
 
 | | Co-crystal structure of MDM2 with inhibitor (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide | | Descriptor: | (2'S,3R,4'S,5'R)-N-(2-aminoethyl)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Huang, X, Gonzalez-Lopez de Turiso, F, Sun, D, Yosup, R, Bartberger, M.D, Beck, H.P, Cannon, J, Shaffer, P, Oliner, J.D, Olson, S.H, Medina, J.C. | | Deposit date: | 2013-03-26 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Rational Design and Binding Mode Duality of MDM2-p53 Inhibitors.
J.Med.Chem., 56, 2013
|
|
