7OHV
 
 | |
3VZL
 
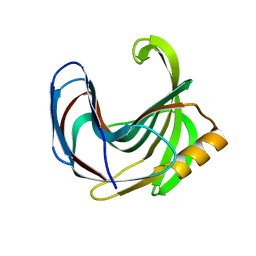 | | Crystal structure of the Bacillus circulans endo-beta-(1,4)-xylanase (BcX) N35H mutant | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Ludwiczek, M.L, D'Angelo, I, Yalloway, G.N, Okon, M, Nielsen, J.E, Strynadka, N.C, Withers, S.G, McIntosh, L.P. | | Deposit date: | 2012-10-15 | | Release date: | 2013-05-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Strategies for modulating the pH-dependent activity of a family 11 glycoside hydrolase
Biochemistry, 52, 2013
|
|
3W01
 
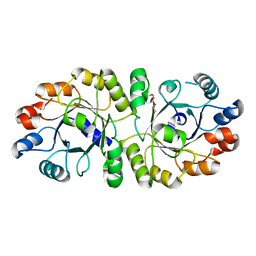 | | Crystal structure of PcrB complexed with PEG from Staphylococcus aureus subsp. aureus Mu3 | | Descriptor: | Heptaprenylglyceryl phosphate synthase, TRIETHYLENE GLYCOL | | Authors: | Ren, F, Feng, X, Ko, T.P, Huang, C.H, Hu, Y, Chan, H.C, Liu, Y.L, Wang, K, Chen, C.C, Pang, X, He, M, Li, Y, Oldfield, E, Guo, R.T. | | Deposit date: | 2012-10-17 | | Release date: | 2012-12-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Insights into TIM-barrel prenyl transferase mechanisms: crystal structures of PcrB from Bacillus subtilis and Staphylococcus aureus
Chembiochem, 14, 2013
|
|
3QL0
 
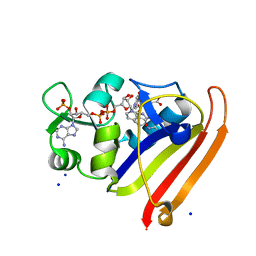 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
7OHX
 
 | |
7OHY
 
 | |
1PK5
 
 | |
8I68
 
 | | Crystal structure of Mycobacterium tuberculosis Uracil-DNA glycosylase in complex with Uric acid, Form III | | Descriptor: | 1,2-ETHANEDIOL, URIC ACID, Uracil-DNA glycosylase | | Authors: | Raj, P, Paul, A, Gopal, B. | | Deposit date: | 2023-01-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structures of non-uracil ring fragments in complex with Mycobacterium tuberculosis uracil DNA glycosylase (MtUng) as a starting point for novel inhibitor design: A case study with the barbituric acid fragment.
Eur.J.Med.Chem., 258, 2023
|
|
7KRR
 
 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7T3M
 
 | | SARS-CoV-2 S (Spike Glycoprotein) D614G with Three (3) RBDs Up, Bound to Antibody 2-7 scFv, composite map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody 2-7 scFv, ... | | Authors: | Byrne, P.O, McLellan, J.S. | | Deposit date: | 2021-12-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | IgG-like bispecific antibodies with potent and synergistic neutralization against circulating SARS-CoV-2 variants of concern.
Nat Commun, 13, 2022
|
|
7KRS
 
 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-24 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
7OHT
 
 | |
5ELW
 
 | | A. thaliana IGPD2 in complex with the triazole-phosphonate inhibitor, (R)-C348, to 1.36A resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Bisson, C, Britton, K.L, Sedelnikova, S.E, Rodgers, H.F, Eadsforth, T.C, Viner, R.C, Hawkes, T.R, Baker, P.J, Rice, D.W. | | Deposit date: | 2015-11-05 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Mirror-Image Packing Provides a Molecular Basis for the Nanomolar Equipotency of Enantiomers of an Experimental Herbicide.
Angew.Chem.Int.Ed.Engl., 55, 2016
|
|
1CNN
 
 | | OMEGA-CONOTOXIN MVIIC FROM CONUS MAGUS | | Descriptor: | OMEGA-CONOTOXIN MVIIC | | Authors: | Nielsen, K.J, Adams, D, Thomas, L, Bond, T, Alewood, P.F, Craik, D.J, Lewis, R.J. | | Deposit date: | 1999-05-20 | | Release date: | 2000-05-31 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure-activity relationships of omega-conotoxins MVIIA, MVIIC and 14 loop splice hybrids at N and P/Q-type calcium channels.
J.Mol.Biol., 289, 1999
|
|
6QQL
 
 | | Crystal structure of Porphyromonas gingivalis glutaminyl cyclase | | Descriptor: | Glutamine cyclotransferase, ZINC ION | | Authors: | Linnert, M, Piechotta, A, Parthier, C, Taudte, N, Kolenko, P, Rahfeld, J, Potempa, J, Stubbs, M.T. | | Deposit date: | 2019-02-18 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | Mammalian-like type II glutaminyl cyclases in Porphyromonas gingivalis and other oral pathogenic bacteria as targets for treatment of periodontitis.
J.Biol.Chem., 296, 2021
|
|
2MVW
 
 | | Solution structure of the TRIM19 B-box1 (B1) of human promyelocytic leukemia (PML) | | Descriptor: | Protein PML, ZINC ION | | Authors: | Huang, S, Naik, M.T, Fan, P, Wang, Y, Chang, C, Huang, T. | | Deposit date: | 2014-10-17 | | Release date: | 2014-11-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The B-box 1 dimer of human promyelocytic leukemia protein.
J.Biomol.Nmr, 60, 2014
|
|
3QRC
 
 | | The crystal structure of Ail, the attachment invasion locus protein of Yersinia pestis, in complex with the heparin analogue sucrose octasulfate | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, Attachment invasion locus protein | | Authors: | Yamashita, S, Lukacik, P, Noinaj, N, Buchanan, S.K. | | Deposit date: | 2011-02-17 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.852 Å) | | Cite: | Structural Insights into Ail-Mediated Adhesion in Yersinia pestis.
Structure, 19, 2011
|
|
2Z43
 
 | | Structure of a twinned crystal of RadA | | Descriptor: | DNA repair and recombination protein radA | | Authors: | Chen, L.T, Ko, T.P, Wang, A.H.J, Wang, T.F. | | Deposit date: | 2007-06-12 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural and functional analyses of five conserved positively charged residues in the L1 and N-terminal DNA binding motifs of archaeal RADA protein
Plos One, 2, 2007
|
|
7OHR
 
 | | Nog1-TAP associated immature ribosomal particle population E from S. cerevisiae | | Descriptor: | 25S rRNA, 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Milkereit, P, Poell, G. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (4.72 Å) | | Cite: | Analysis of subunit folding contribution of three yeast large ribosomal subunit proteins required for stabilisation and processing of intermediate nuclear rRNA precursors.
Plos One, 16, 2021
|
|
7KRQ
 
 | | Structural impact on SARS-CoV-2 spike protein by D614G substitution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zhang, J, Cai, Y.F, Xiao, T.S, Lu, J.M, Peng, H.Q, Sterling, S.M, Walsh Jr, R.M, Volloch, S.R, Zhu, H.S, Woosley, A.N, Yang, W, Sliz, P, Chen, B. | | Deposit date: | 2020-11-20 | | Release date: | 2021-03-31 | | Last modified: | 2021-05-12 | | Method: | ELECTRON MICROSCOPY (3.44 Å) | | Cite: | Structural impact on SARS-CoV-2 spike protein by D614G substitution.
Science, 372, 2021
|
|
1D85
 
 | | STRUCTURAL CONSEQUENCES OF A CARCINOGENIC ALKYLATION LESION ON DNA: EFFECT OF O6-ETHYL-GUANINE ON THE MOLECULAR STRUCTURE OF D(CGC[E6G]AATTCGCG)-NETROPSIN COMPLEX | | Descriptor: | DNA (5'-D(*CP*GP*CP*(G36)P*AP*AP*TP*TP*CP*GP*CP*G)-3'), NETROPSIN | | Authors: | Sriram, M, Van Der Marel, G.A, Roelen, H.L.P.F, Van Boom, J.H, Wang, A.H.-J. | | Deposit date: | 1992-08-24 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural consequences of a carcinogenic alkylation lesion on DNA: effect of O6-ethylguanine on the molecular structure of the d(CGC[e6G]AATTCGCG)-netropsin complex.
Biochemistry, 31, 1992
|
|
3QTC
 
 | | Crystal structure of the catalytic domain of MmOmeRS, an O-methyl tyrosyl-tRNA synthetase evolved from Methanosarcina mazei PylRS, complexed with O-methyl tyrosine and AMP-PNP | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, O-methyl-L-tyrosine, ... | | Authors: | Dellas, N, Takimoto, J.K, Noel, J.P, Wang, L. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Stereochemical Basis for Engineered Pyrrolysyl-tRNA Synthetase and the Efficient in Vivo Incorporation of Structurally Divergent Non-native Amino Acids.
Acs Chem.Biol., 6, 2011
|
|
1PFV
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH DIFLUOROMETHIONINE | | Descriptor: | Methionyl-tRNA synthetase, S-(DIFLUOROMETHYL)HOMOCYSTEINE, ZINC ION | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
1PG2
 
 | | METHIONYL-TRNA SYNTHETASE FROM ESCHERICHIA COLI COMPLEXED WITH METHIONINE AND ADENOSINE | | Descriptor: | ADENOSINE, METHIONINE, Methionyl-tRNA synthetase | | Authors: | Crepin, T, Schmitt, E, Mechulam, Y, Sampson, P.B, Vaughan, M.D, Honek, J.F, Blanquet, S. | | Deposit date: | 2003-05-27 | | Release date: | 2004-02-17 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Use of analogues of methionine and methionyl adenylate to sample conformational changes during catalysis in Escherichia coli methionyl-tRNA synthetase.
J.Mol.Biol., 332, 2003
|
|
7T7L
 
 | | Structure of human G9a SET-domain (EHMT2) in complex with covalent inhibitor (Compound 1) | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)prop-2-enamide, N-(6-methoxy-4-{[1-(propan-2-yl)piperidin-4-yl]amino}-7-[3-(pyrrolidin-1-yl)propoxy]quinazolin-2-yl)propanamide, ... | | Authors: | Park, K.-S, Kumar, P. | | Deposit date: | 2021-12-15 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of the First-in-Class G9a/GLP Covalent Inhibitors.
J.Med.Chem., 65, 2022
|
|
