8GUA
 
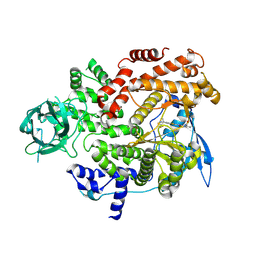 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant E542K in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-12-07 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6SAV
 
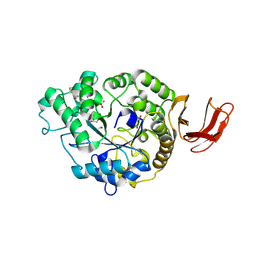 | | Structural and functional characterisation of three novel fungal amylases with enhanced stability and pH tolerance | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Alpha-amylase, CALCIUM ION, ... | | Authors: | Roth, C, Moroz, O.V, Turkenburg, J.P, Blagova, E, Waterman, J, Ariza, A, Ming, L, Tianqi, S, Andersen, C, Davies, G.J, Wilson, K.S. | | Deposit date: | 2019-07-17 | | Release date: | 2019-10-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and Functional Characterization of Three Novel Fungal Amylases with Enhanced Stability and pH Tolerance.
Int J Mol Sci, 20, 2019
|
|
6SC2
 
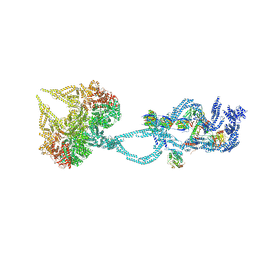 | | Structure of the dynein-2 complex; IFT-train bound model | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Cytoplasmic dynein 2 light intermediate chain 1, ... | | Authors: | Toropova, K, Zalyte, R, Mukhopadhyay, A.G, Mladenov, M, Carter, A.P, Roberts, A.J. | | Deposit date: | 2019-07-23 | | Release date: | 2019-09-04 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of the dynein-2 complex and its assembly with intraflagellar transport trains.
Nat.Struct.Mol.Biol., 26, 2019
|
|
5J93
 
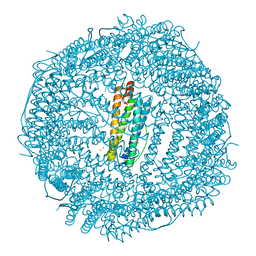 | | Five minutes iron loaded Rana Catesbeiana H' ferritin variant E57A/E136A/D140A | | Descriptor: | CHLORIDE ION, FE (II) ION, Ferritin, ... | | Authors: | Pozzi, C, Di Pisa, F, Mangani, S, Bernacchioni, C, Turano, P. | | Deposit date: | 2016-04-08 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Ferroxidase Activity in Eukaryotic Ferritin is Controlled by Accessory-Iron-Binding Sites in the Catalytic Cavity.
Chemistry, 22, 2016
|
|
5XZD
 
 | |
4QG8
 
 | | crystal structure of PKM2-K305Q mutant | | Descriptor: | GLYCEROL, MAGNESIUM ION, MALONATE ION, ... | | Authors: | Wang, P, Sun, C, Zhu, T, Xu, Y. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insight into mechanisms for dynamic regulation of PKM2.
Protein Cell, 6, 2015
|
|
1BRI
 
 | | BARNASE MUTANT WITH ILE 76 REPLACED BY ALA | | Descriptor: | BARNASE | | Authors: | Cramer, P.C, Buckle, A, Fersht, A. | | Deposit date: | 1995-03-09 | | Release date: | 1995-07-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and energetic responses to cavity-creating mutations in hydrophobic cores: observation of a buried water molecule and the hydrophilic nature of such hydrophobic cavities.
Biochemistry, 35, 1996
|
|
6J0E
 
 | | Structures of two ArsR As(III)-responsive repressors: implications for the mechanism of derepression | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ARSENIC, Arsenic responsive repressor ArsR | | Authors: | Prabaharan, C, Kandavelu, P, Packianathan, C, Rosen, P.B, Thiyagarajan, S. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two ArsR As(III)-responsive transcriptional repressors: Implications for the mechanism of derepression.
J.Struct.Biol., 207, 2019
|
|
4QHI
 
 | | Crystal structure of Methanocaldococcus jannaschii selecase mutant R36W | | Descriptor: | CHLORIDE ION, GLYCEROL, Uncharacterized protein MJ1213, ... | | Authors: | Lopez-pelegrin, M, Cerda-costa, N, Cintas-pedrola, A, Herranz-trillo, F, Bernado, P, Peinado, J.R, Arolas, J.L, Gomis-ruth, F.X. | | Deposit date: | 2014-05-28 | | Release date: | 2014-07-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Multiple stable conformations account for reversible concentration-dependent oligomerization and autoinhibition of a metamorphic metallopeptidase
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1OZ1
 
 | | P38 MITOGEN-ACTIVATED KINASE IN COMPLEX WITH 4-AZAINDOLE INHIBITOR | | Descriptor: | 3-(4-FLUOROPHENYL)-2-PYRIDIN-4-YL-1H-PYRROLO[3,2-B]PYRIDIN-1-OL, Mitogen-activated protein kinase 14 | | Authors: | Lovejoy, B, Villasenor, A, Browner, M, Dunten, P. | | Deposit date: | 2003-04-07 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Design and synthesis of 4-azaindoles as inhibitors of p38 MAP kinase.
J.Med.Chem., 46, 2003
|
|
4L19
 
 | | Matrix metalloproteinase-13 complexed with selective inhibitor compound Q1 | | Descriptor: | 2-[(4-methylbenzyl)sulfanyl]-3,5,6,7-tetrahydro-4H-cyclopenta[d]pyrimidin-4-one, CALCIUM ION, Collagenase 3, ... | | Authors: | Minond, D, Spicer, T.P, Jiang, J, Taylor, A.B, Hart, P.J, Roush, W.R, Fields, G.B, Hodder, P.S. | | Deposit date: | 2013-06-02 | | Release date: | 2014-12-10 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Characterization of selective exosite-binding inhibitors of matrix metalloproteinase 13 that prevent articular cartilage degradation in vitro.
J.Med.Chem., 57, 2014
|
|
5IUA
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12b at 2.2A resolution | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(furan-2-yl)-N~5~-[3-(4-phenylpiperazin-1-yl)propyl][1,2,4]triazolo[1,5-a][1,3,5]triazine-5,7-diamine, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
5EZS
 
 | |
1F69
 
 | |
5IU8
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with compound 12f at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-(furan-2-yl)-N~5~-[2-(4-methylpiperazin-1-yl)ethyl][1,2,4]triazolo[1,5-a][1,3,5]triazine-5,7-diamine, ... | | Authors: | Segala, E, Guo, D, Cheng, R.K.Y, Bortolato, A, Deflorian, F, Dore, A.S, Errey, J.C, Heitman, L.H, Ijzerman, A.P, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2016-03-17 | | Release date: | 2016-06-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Controlling the Dissociation of Ligands from the Adenosine A2A Receptor through Modulation of Salt Bridge Strength.
J.Med.Chem., 59, 2016
|
|
5M1W
 
 | | Structure of a stable G-hairpin | | Descriptor: | DNA (5'-D(*GP*TP*GP*TP*GP*GP*GP*TP*GP*TP*G)-3') | | Authors: | Gajarsky, M, Zivkovic, M.L, Stadlbauer, P, Pagano, B, Fiala, R, Amato, J, Tomaska, L, Sponer, J, Plavec, J, Trantirek, L. | | Deposit date: | 2016-10-11 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of a Stable G-Hairpin.
J. Am. Chem. Soc., 139, 2017
|
|
8G5E
 
 | | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535 | | Descriptor: | Histone-lysine N-methyltransferase SETDB1, N~4~-[6-(dimethylamino)hexyl]-N~2~-[5-(dimethylamino)pentyl]-6,7-dimethoxyquinazoline-2,4-diamine, UNKNOWN ATOM OR ION | | Authors: | Beldar, S, Dong, A, Brown, P.J, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-02-13 | | Release date: | 2023-02-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Crystal Structure of SETDB1 Tudor domain in complex with UNC6535
To be published
|
|
1X09
 
 | | Crystal structure of the D26A mutant UPPs in complex with magnesium and isopentenyl pyrophosphate | | Descriptor: | 3-METHYLBUT-3-ENYL TRIHYDROGEN DIPHOSPHATE, MAGNESIUM ION, Undecaprenyl pyrophosphate synthetase | | Authors: | Guo, R.-T, Ko, T.-P, Chen, A.P.-C, Kuo, C.-J, Wang, A.H.-J, Liang, P.-H. | | Deposit date: | 2005-03-15 | | Release date: | 2005-03-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structures of undecaprenyl pyrophosphate synthase in complex with magnesium, isopentenyl pyrophosphate, and farnesyl thiopyrophosphate: roles of the metal ion and conserved residues in catalysis.
J.Biol.Chem., 280, 2005
|
|
5F0C
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium tuberculosis in complex with compound 4 at 1.87A resolution | | Descriptor: | 3-cyclopentyl-~{N}-(3-piperidin-1-ylphenyl)propanamide, HTH-type transcriptional regulator EthR, SULFATE ION | | Authors: | Surade, S, Blaszczyk, M, Nikiforov, P.O, Abell, C, Blundell, T.L. | | Deposit date: | 2015-11-27 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | A fragment merging approach towards the development of small molecule inhibitors of Mycobacterium tuberculosis EthR for use as ethionamide boosters.
Org.Biomol.Chem., 14, 2016
|
|
5EX2
 
 | |
6ETS
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 1 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.333 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6SCQ
 
 | |
6SCX
 
 | | Crystal structure of the catalytic domain of human NUDT12 in complex with 7-methyl-guanosine-5'-triphosphate | | Descriptor: | 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE, CADMIUM ION, Peroxisomal NADH pyrophosphatase NUDT12 | | Authors: | McCarthy, A.A, Chen, K.M, Wu, H, Li, L, Homolka, D, Gos, P, Fleury-Olela, F, Pillai, R.S. | | Deposit date: | 2019-07-25 | | Release date: | 2020-01-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Decapping Enzyme NUDT12 Partners with BLMH for Cytoplasmic Surveillance of NAD-Capped RNAs.
Cell Rep, 29, 2019
|
|
6J76
 
 | | Structure of 3,6-anhydro-L-galactose Dehydrogenase in Complex with NAP | | Descriptor: | Aldehyde dehydrogenase A, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Li, P.Y, Wang, Y, Chen, X.L, Zhang, Y.Z. | | Deposit date: | 2019-01-17 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.368 Å) | | Cite: | 3,6-Anhydro-L-Galactose Dehydrogenase VvAHGD is a Member of a New Aldehyde Dehydrogenase Family and Catalyzes by a Novel Mechanism with Conformational Switch of Two Catalytic Residues Cysteine 282 and Glutamate 248.
J.Mol.Biol., 432, 2020
|
|
4Q1J
 
 | | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-branching | | Descriptor: | 1,2-ETHANEDIOL, Polyketide biosynthesis enoyl-CoA isomerase PksI, SODIUM ION | | Authors: | Nair, A.V, Race, P.R, Till, M. | | Deposit date: | 2014-04-03 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure and mechanism of a dehydratase/decarboxylase enzyme couple involved in polyketide beta-methyl branch incorporation.
Sci Rep, 10, 2020
|
|
