7AWQ
 
 | | Structure of the thermostabilized EAAT1 cryst-E386Q mutant in complex with L-ASP, sodium ions and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, ASPARTIC ACID, BARIUM ION, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
7AWN
 
 | | Structure of the thermostabilized EAAT1 cryst mutant in complex with rubidium and barium and the allosteric inhibitor UCPH101 | | Descriptor: | 2-Amino-5,6,7,8-tetrahydro-4-(4-methoxyphenyl)-7-(naphthalen-1-yl)-5-oxo-4H-chromene-3-carbonitrile, BARIUM ION, Excitatory amino acid transporter 1,Neutral amino acid transporter B(0),Excitatory amino acid transporter 1, ... | | Authors: | Canul-Tec, J.C, Legrand, P, Reyes, N. | | Deposit date: | 2020-11-08 | | Release date: | 2021-10-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.92 Å) | | Cite: | The ion-coupling mechanism of human excitatory amino acid transporters.
Embo J., 41, 2022
|
|
1GC1
 
 | | HIV-1 GP120 CORE COMPLEXED WITH CD4 AND A NEUTRALIZING HUMAN ANTIBODY | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ANTIBODY 17B, CD4, ... | | Authors: | Kwong, P.D, Wyatt, R, Robinson, J, Sweet, R.W, Sodroski, J, Hendrickson, W.A. | | Deposit date: | 1998-06-15 | | Release date: | 1998-07-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody.
Nature, 393, 1998
|
|
4WH0
 
 | | YcaC from Pseudomonas aeruginosa with S-mercaptocysteine active site cysteine | | Descriptor: | CHLORIDE ION, Putative hydrolase | | Authors: | Groftehauge, M.K, Truan, D, Vasil, A, Denny, P.W, Vasil, M.L, Pohl, E. | | Deposit date: | 2014-09-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.563 Å) | | Cite: | Crystal Structure of a Hidden Protein, YcaC, a Putative Cysteine Hydrolase from Pseudomonas aeruginosa, with and without an Acrylamide Adduct.
Int J Mol Sci, 16, 2015
|
|
6UQ8
 
 | |
5TBJ
 
 | | Crystal structure of mouse CARM1 in complex with inhibitor LH1452 | | Descriptor: | 1,2-DIMETHOXYETHANE, 1,2-ETHANEDIOL, 1-(2-METHOXY-ETHOXY)-2-{2-[2-(2-METHOXY-ETHOXY]-ETHOXY}-ETHANE, ... | | Authors: | Cura, V, Marechal, N, Troffer-Charlier, N, Halby, L, Arimondo, P, Bonnefond, L, Cavarelli, J. | | Deposit date: | 2016-09-12 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Hijacking DNA methyltransferase transition state analogues to produce chemical scaffolds for PRMT inhibitors.
Philos.Trans.R.Soc.Lond.B Biol.Sci., 373, 2018
|
|
7AQL
 
 | |
6B45
 
 | | Cryo-EM structure of Type I-F CRISPR crRNA-guided Csy surveillance complex | | Descriptor: | CRISPR-associated endonuclease Cas6/Csy4, CRISPR-associated protein Csy1, CRISPR-associated protein Csy2, ... | | Authors: | Guo, T.W, Bartesaghi, A, Yang, H, Falconieri, V, Rao, P, Merk, A, Fox, T, Earl, L, Patel, D.J, Subramaniam, S. | | Deposit date: | 2017-09-25 | | Release date: | 2017-10-18 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM Structures Reveal Mechanism and Inhibition of DNA Targeting by a CRISPR-Cas Surveillance Complex.
Cell, 171, 2017
|
|
1WAP
 
 | |
7T86
 
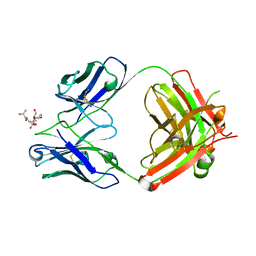 | | Crystal Structure of Fab CR5133 / Phospho-SD Peptide Complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Antibody Fab CR5133 Heavy Chain, Antibody Fab CR5133 Light Chain, ... | | Authors: | Luo, J, Malia, T.J, Buckley, P.T. | | Deposit date: | 2021-12-15 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multivalent human antibody-centyrin fusion protein to prevent and treat Staphylococcus aureus infections.
Cell Host Microbe, 31, 2023
|
|
6OMY
 
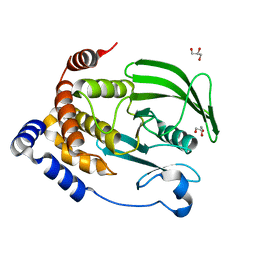 | | Protein Tyrosine Phosphatase 1B (1-301), P180A mutant, apo state | | Descriptor: | GLYCEROL, Tyrosine-protein phosphatase non-receptor type 1 | | Authors: | Cui, D.S, Lipchock, J.M, Loria, J.P. | | Deposit date: | 2019-04-19 | | Release date: | 2019-08-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Uncovering the Molecular Interactions in the Catalytic Loop That Modulate the Conformational Dynamics in Protein Tyrosine Phosphatase 1B.
J.Am.Chem.Soc., 141, 2019
|
|
6UX5
 
 | |
8D9P
 
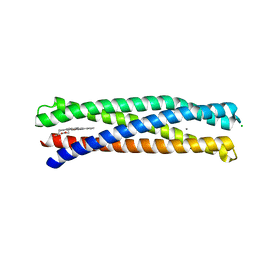 | | De Novo Photosynthetic Reaction Center Protein Equipped with Heme B and Mn(II) cations | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Ennist, N.M, Stayrook, S.E, Dutton, P.L, Moser, C.C. | | Deposit date: | 2022-06-10 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Rational design of photosynthetic reaction center protein maquettes.
Front Mol Biosci, 9, 2022
|
|
6V1D
 
 | | Crystal structure of human trefoil factor 1 | | Descriptor: | Trefoil factor 1 | | Authors: | Jarva, M.A, Lingford, J.P, John, A, Scott, N.E, Goddard-Borger, E.D. | | Deposit date: | 2019-11-20 | | Release date: | 2019-12-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Trefoil factors share a lectin activity that defines their role in mucus.
Nat Commun, 11, 2020
|
|
1FY1
 
 | | [R23S,F25E]HBP, A MUTANT OF HUMAN HEPARIN BINDING PROTEIN (CAP37) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ETHANOL, ... | | Authors: | Kastrup, J.S, Linde, V, Pedersen, A.K, Stoffer, B, Iversen, L.F, Larsen, I.K, Rasmussen, P.B, Flodgaard, H.J, Bjorn, S.E. | | Deposit date: | 2000-09-28 | | Release date: | 2001-09-28 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Two mutants of human heparin binding protein (CAP37): toward the understanding of the nature of lipid A/LPS and BPTI binding.
Proteins, 42, 2001
|
|
1G1A
 
 | | THE CRYSTAL STRUCTURE OF DTDP-D-GLUCOSE 4,6-DEHYDRATASE (RMLB)FROM SALMONELLA ENTERICA SEROVAR TYPHIMURIUM | | Descriptor: | DTDP-D-GLUCOSE 4,6-DEHYDRATASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Allard, S.T.M, Giraud, M.-F, Whitfield, C, Graninger, M, Messner, P, Naismith, J.H. | | Deposit date: | 2000-10-11 | | Release date: | 2001-03-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | The crystal structure of dTDP-D-Glucose 4,6-dehydratase (RmlB) from Salmonella enterica serovar Typhimurium, the second enzyme in the dTDP-l-rhamnose pathway.
J.Mol.Biol., 307, 2001
|
|
4WVI
 
 | | Crystal structure of the Type-I signal peptidase from Staphylococcus aureus (SpsB) in complex with a substrate peptide (pep2). | | Descriptor: | Maltose-binding periplasmic protein,Signal peptidase IB, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, substrate peptide (pep2) | | Authors: | Young, P.G, Ting, Y.T, Baker, E.N. | | Deposit date: | 2014-11-05 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Peptide binding to a bacterial signal peptidase visualized by peptide tethering and carrier-driven crystallization.
IUCrJ, 3, 2016
|
|
6Q43
 
 | | Atomic resolution crystal structure of an ABA collagen heterotrimer | | Descriptor: | GLYCEROL, Leading Chain of the ABA collagen heterotrimer, Middle Chain of the ABA collagen heterotrimer, ... | | Authors: | Jalan, A.A, Hartgerink, J.D, Brear, P, Leitinger, B, Farndale, R.W. | | Deposit date: | 2018-12-05 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Atomic resolution crystal structure of an AAB collagen heterotrimer
Nat.Chem.Biol., 2019
|
|
1G2R
 
 | | Structure of Cytosolic Protein of Unknown Function Coded by Gene from NUSA/INFB Region, a YlxR Homologue | | Descriptor: | HYPOTHETICAL CYTOSOLIC PROTEIN, SULFATE ION | | Authors: | Osipiuk, J, Gornicki, P, Maj, L, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2000-10-20 | | Release date: | 2001-08-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Streptococcus pneumonia YlxR at 1.35 A shows a putative new fold.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
7EV0
 
 | | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Singh, J, Ahmad, M.I, Maurya, A, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2021-05-19 | | Release date: | 2021-06-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of pepsin cleaved C-terminal half of lactoferrin at 2.7A resolution
To Be Published
|
|
5T6D
 
 | | 2.10 A resolution structure of Norovirus 3CL protease in complex with the dipeptidyl inhibitor 7l (hexagonal form) | | Descriptor: | 3-cyclohexyl-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-N~2~-{[3-(4-methoxyphenoxy)propyl]sulfonyl}-L- alaninamide, Genome polyprotein | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Rathnayake, A.D, Damalanka, V.C, Weerawarna, P.M, Doyle, S.T, Alsoudi, A.F, Dissanayake, D.M.P, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2016-09-01 | | Release date: | 2016-11-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-based exploration and exploitation of the S4 subsite of norovirus 3CL protease in the design of potent and permeable inhibitors.
Eur J Med Chem, 126, 2016
|
|
1JQ8
 
 | | Design of specific inhibitors of phospholipase A2: Crystal structure of a complex formed between phospholipase A2 from Daboia russelli pulchella and a designed pentapeptide Leu-Ala-Ile-Tyr-Ser at 2.0 resolution | | Descriptor: | ACETIC ACID, Peptide inhibitor, Phospholipase A2, ... | | Authors: | Chandra, V, Jasti, J, Kaur, P, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2001-08-04 | | Release date: | 2002-11-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design of specific peptide inhibitors of phospholipase A2: structure of a complex formed between Russell's viper phospholipase A2 and a designed peptide Leu-Ala-Ile-Tyr-Ser (LAIYS).
ACTA CRYSTALLOGR.,SECT.D, 58, 2002
|
|
7B3H
 
 | | Notum complex with ARUK3003909 | | Descriptor: | 1,2-ETHANEDIOL, 6-((3-(trifluoromethoxy)phenyl)thio)-[1,2,4]triazolo[4,3-b]pyridazin-3(2H)-one, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y, Fish, P. | | Deposit date: | 2020-11-30 | | Release date: | 2021-12-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Virtual Screening Directly Identifies New Fragment-Sized Inhibitors of Carboxylesterase Notum with Nanomolar Activity.
J.Med.Chem., 65, 2022
|
|
1GGW
 
 | |
6UUN
 
 | | CryoEM Structure of the active Adrenomedullin 1 receptor G protein complex with adrenomedullin peptide | | Descriptor: | ADM, Calcitonin gene-related peptide type 1 receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Belousoff, M.J, Liang, Y.L, Sexton, P, Danev, R. | | Deposit date: | 2019-10-30 | | Release date: | 2020-03-25 | | Last modified: | 2020-04-29 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure and Dynamics of Adrenomedullin Receptors AM1and AM2Reveal Key Mechanisms in the Control of Receptor Phenotype by Receptor Activity-Modifying Proteins.
Acs Pharmacol Transl Sci, 3, 2020
|
|
