6EZZ
 
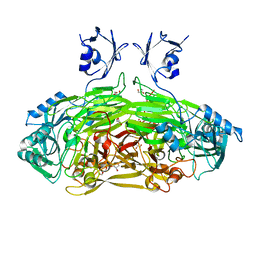 | | Crystal structure of Escherichia coli amine oxidase mutant E573Q | | Descriptor: | CALCIUM ION, COPPER (II) ION, GLYCEROL, ... | | Authors: | Gaule, T.G, Smith, M.A, Tych, K.M, Pirrat, P, Trinh, C.H, Pearson, A.R, Knowles, P.F, McPherson, M.J. | | Deposit date: | 2017-11-16 | | Release date: | 2018-08-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Oxygen Activation Switch in the Copper Amine Oxidase of Escherichia coli.
Biochemistry, 57, 2018
|
|
5FZ3
 
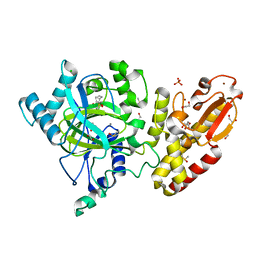 | | Crystal structure of the catalytic domain of human JARID1B in complex with Maybridge fragment 3,6-Dihydroxybenzonorbornane (N08776b) (ligand modelled based on PANDDA event map) | | Descriptor: | (1R,4S)-1,2,3,4-tetrahydro-1,4-methanonaphthalene-5,8-diol, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Nowak, R, Krojer, T, Johansson, C, Gileadi, C, Kupinska, K, Strain-Damerell, C, Szykowska, A, Pearce, N, Talon, R, Collins, P, Burgess-Brown, N.A, Arrowsmith, C.H, Bountra, C, Edwards, A.M, von Delft, F, Brennan, P.E, Oppermann, U. | | Deposit date: | 2016-03-10 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Catalytic Domain of Human Jarid1B in Complex with N08776B
To be Published
|
|
5Y98
 
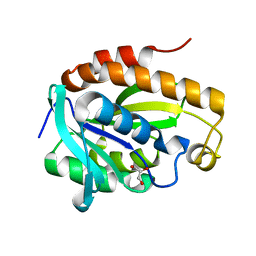 | | Crystal structure of native unbound peptidyl tRNA hydrolase from Acinetobacter baumannii at 1.36 A resolution | | Descriptor: | GLYCEROL, Peptidyl-tRNA hydrolase | | Authors: | Iqbal, N, Singh, N, Kaushik, S, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-23 | | Release date: | 2017-09-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Search of multiple hot spots on the surface of peptidyl-tRNA hydrolase: structural, binding and antibacterial studies.
Biochem. J., 475, 2018
|
|
5AJX
 
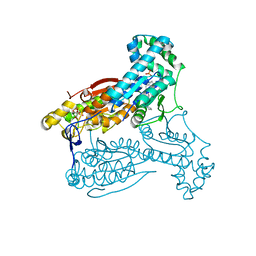 | | Human PFKFB3 in complex with an indole inhibitor 3 | | Descriptor: | (2S)-N-[4-[3-cyano-1-[(3,5-dimethyl-1,2-oxazol-4-yl)methyl]indol-5-yl]oxyphenyl]pyrrolidine-2-carboxamide, 6-O-phosphono-beta-D-fructofuranose, 6-PHOSPHOFRUCTO-2-KINASE/FRUCTOSE-2,6-BISPHOSPHATASE 3, ... | | Authors: | Boyd, S, Brookfield, J.L, Critchlow, S.E, Cumming, I.A, Curtis, N.J, Debreczeni, J.E, Degorce, S.L, Donald, C, Evans, N.J, Groombridge, S, Hopcroft, P, Jones, N.P, Kettle, J.G, Lamont, S, Lewis, H.J, MacFaull, P, McLoughlin, S.B, Rigoreau, L.J.M, Smith, J.M, St-Gallay, S, Stock, J.K, Wheatley, E.R, Winter, J, Wingfield, J. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure-Based Design of Potent and Selective Inhibitors of the Metabolic Kinase Pfkfb3.
J.Med.Chem., 58, 2015
|
|
6H2D
 
 | |
5FSP
 
 | | Structure of thermolysin prepared by the 'soak-and-freeze' method under 100 bar of krypton pressure | | Descriptor: | CALCIUM ION, KRYPTON, LYSINE, ... | | Authors: | Lafumat, B, Mueller-Dieckmann, C, Colloc'h, N, Prange, T, Royant, A, van der Linden, p, Carpentier, P. | | Deposit date: | 2016-01-06 | | Release date: | 2016-10-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Gas-Sensitive Biological Crystals Processed in Pressurized Oxygen and Krypton Atmospheres: Deciphering Gas Channels in Proteins Using a Novel `Soak-and-Freeze' Methodology.
J.Appl.Crystallogr., 49, 2016
|
|
7PQF
 
 | | Crystal structure of Campylobacter jejuni DsbA2 | | Descriptor: | Thiol:disulfide interchange protein DsbA/DsbL | | Authors: | Wilk, P, Banas, A.M, Bocian-Ostrzycka, K.M, Jagusztyn-Krynicka, E.K. | | Deposit date: | 2021-09-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Interplay between DsbA1, DsbA2 and C8J_1298 Periplasmic Oxidoreductases of Campylobacter jejuni and Their Impact on Bacterial Physiology and Pathogenesis.
Int J Mol Sci, 22, 2021
|
|
5FH7
 
 | | Crystal structure of the fifth bromodomain of human PB1 in complex with compound 18 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloranyl-3-[(dimethylamino)methyl]-4~{H}-pyrrolo[1,2-a]quinazolin-5-one, Protein polybromo-1 | | Authors: | Tallant, C, Sutherell, C.L, Siejka, P, Sorrell, F.J, Krojer, T, Picaud, S, Fedorov, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Brennan, P.E, Ley, S.V, Knapp, S. | | Deposit date: | 2015-12-21 | | Release date: | 2016-06-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Identification and Development of 2,3-Dihydropyrrolo[1,2-a]quinazolin-5(1H)-one Inhibitors Targeting Bromodomains within the Switch/Sucrose Nonfermenting Complex.
J.Med.Chem., 59, 2016
|
|
7UQB
 
 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1-D52A strain with AlF4 | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-19 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
8POL
 
 | | Crystal structure of Plasmodium falciparum Sub1 protease | | Descriptor: | CALCIUM ION, PHOSPHATE ION, Subtilisin-like protease 1 | | Authors: | Martinez, M, Bouillon, A, Haouz, A, Barale, J.C, Alzari, P.M. | | Deposit date: | 2023-07-05 | | Release date: | 2024-03-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Prodomain-driven enzyme dimerization: a pH-dependent autoinhibition mechanism that controls Plasmodium Sub1 activity before merozoite egress.
Mbio, 15, 2024
|
|
7ZKC
 
 | | Catalytic domain of UDP-Glucose Glycoprotein Glucosyltransferase from Chaetomium thermophilum (apo form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, UDP-glucose-glycoprotein glucosyltransferase-like protein | | Authors: | Roversi, P, Zitzmann, N, Bayo, Y, Le Cornu, J.D. | | Deposit date: | 2022-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.769 Å) | | Cite: | A quinolin-8-ol sub-millimolar inhibitor of UGGT, the ER glycoprotein folding quality control checkpoint.
Iscience, 26, 2023
|
|
8OZ8
 
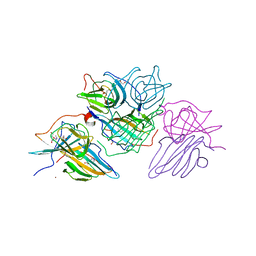 | | Crystal Structure of an Hydroxynitrile lyase variant (H96A) from Granulicella tundricola | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 3-CARBOXY-N,N,N-TRIMETHYLPROPAN-1-AMINIUM, BROMIDE ION, ... | | Authors: | Bento, I, Coloma, J, Hagedoorn, P.-L, Hanefeld, U. | | Deposit date: | 2023-05-08 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Can a Hydroxynitrile Lyase Catalyze an Oxidative Cleavage?
Acs Catalysis, 13, 2023
|
|
5IK9
 
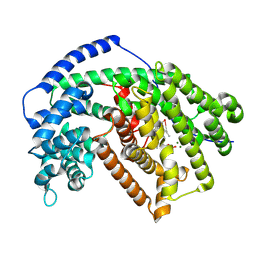 | | Tobacco 5-epi-aristolochene with farnesyl monophosphate | | Descriptor: | (2E,6Z)-3,7,11-trimethyldodeca-2,6,10-trien-1-yl dihydrogen phosphate, 5-epi-aristolochene synthase, MAGNESIUM ION | | Authors: | Koo, H.J, Xu, Y, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
6ARJ
 
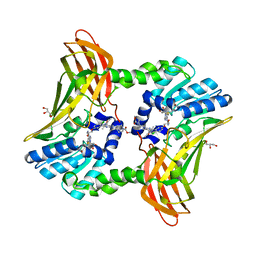 | | Crystal structure of CARM1 with EPZ022302 and SAH | | Descriptor: | GLYCEROL, Histone-arginine methyltransferase CARM1, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Boriack-Sjodin, P.A, Jin, L. | | Deposit date: | 2017-08-22 | | Release date: | 2018-02-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Identification of a CARM1 Inhibitor with Potent In Vitro and In Vivo Activity in Preclinical Models of Multiple Myeloma.
Sci Rep, 7, 2017
|
|
7UOO
 
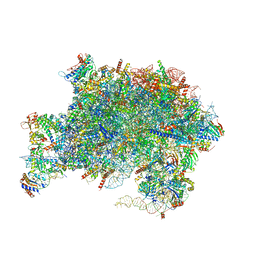 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-13 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.34 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
6SH9
 
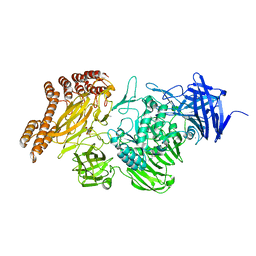 | | EngBF DARPin Fusion 4b D12 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-alpha-N-acetylgalactosaminidase,DARPin 4b D12, ... | | Authors: | Ernst, P, Pluckthun, A, Mittl, P.R.E. | | Deposit date: | 2019-08-06 | | Release date: | 2019-11-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural analysis of biological targets by host:guest crystal lattice engineering.
Sci Rep, 9, 2019
|
|
7UQZ
 
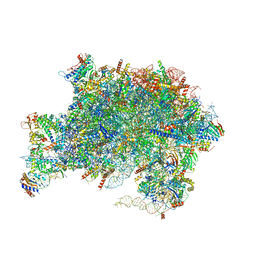 | | Nucleoplasmic pre-60S intermediate of the Nog2 containing pre-rotation state from a SPB1 D52A strain | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 25S rRNA, 5.8S rRNA, ... | | Authors: | Sekulski, K, Cruz, V.E, Weirich, C.S, Erzberger, J.P. | | Deposit date: | 2022-04-20 | | Release date: | 2023-03-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | rRNA methylation by Spb1 regulates the GTPase activity of Nog2 during 60S ribosomal subunit assembly.
Nat Commun, 14, 2023
|
|
6EU9
 
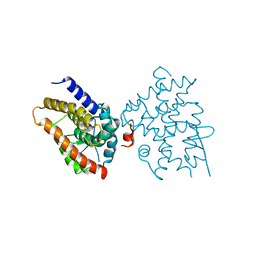 | | Crystal structure of Platynereis dumerilii RAR ligand-binding domain in complex with all-trans retinoic acid | | Descriptor: | RETINOIC ACID, Retinoic acid receptor | | Authors: | Handberg-Thorsager, M, Gutierrez-Mazariegos, J, Arold, S.T, Nadendla, E.K, Bertucci, P.Y, Germain, P, Tomancak, P, Pierzchalski, K, Jones, J.W, Albalat, R, Kane, M.A, Bourguet, W, Laudet, V, Arendt, D, Schubert, M. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | The ancestral retinoic acid receptor was a low-affinity sensor triggering neuronal differentiation.
Sci Adv, 4, 2018
|
|
6EW9
 
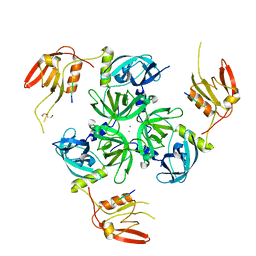 | | CRYSTAL STRUCTURE OF DEGS STRESS SENSOR PROTEASE IN COMPLEX WITH ACTIVATING DNRLGLVYQF PEPTIDE | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, DNRLGLVYQF PEPTIDE, ... | | Authors: | Vetter, I.R, Porfetye, A.T, Stege, P. | | Deposit date: | 2017-11-03 | | Release date: | 2018-04-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of Noncatalytic Lysine Residues from Allosteric Circuits via Covalent Probes.
ACS Chem. Biol., 13, 2018
|
|
6SA6
 
 | | DARPin-Armadillo fusion A5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DARPin-Armadillo fusion A5 | | Authors: | Ernst, P, Honegger, A, van der Valk, F, Ewald, C, Mittl, P.R.E, Pluckthun, A. | | Deposit date: | 2019-07-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rigid fusions of designed helical repeat binding proteins efficiently protect a binding surface from crystal contacts.
Sci Rep, 9, 2019
|
|
7PXQ
 
 | | GH115 alpha-1,2-glucuronidase D303A | | Descriptor: | CALCIUM ION, xylan alpha-1,2-glucuronidase | | Authors: | Wilkens, C, Morth, J.P, Polikarpov, I. | | Deposit date: | 2021-10-08 | | Release date: | 2022-01-19 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A GH115 alpha-glucuronidase structure reveals dimerization-mediated substrate binding and a proton wire potentially important for catalysis.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6B0V
 
 | | Crystal Structure of small molecule ARS-107 covalently bound to K-Ras G12C | | Descriptor: | 1-[3-(4-{[(4,5-dichloro-2-hydroxyphenyl)amino]acetyl}piperazin-1-yl)azetidin-1-yl]propan-1-one, CALCIUM ION, GTPase KRas, ... | | Authors: | Hansen, R, Peters, U, Babbar, A, Chen, Y, Feng, J, Janes, M.R, Li, L.-S, Ren, P, Liu, Y, Zarrinkar, P.P. | | Deposit date: | 2017-09-15 | | Release date: | 2018-05-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | The reactivity-driven biochemical mechanism of covalent KRASG12Cinhibitors.
Nat. Struct. Mol. Biol., 25, 2018
|
|
5IPI
 
 | | Structure of Adeno-associated virus type 2 VLP | | Descriptor: | Capsid protein VP1 | | Authors: | Drouin, L.M, Lins, B, Janssen, M.E, Bennet, A, Chipman, P, McKenna, R, Chen, W, Muzyczka, N, Cardone, G, Baker, T.S, Agbandje-McKenna, M. | | Deposit date: | 2016-03-09 | | Release date: | 2016-07-20 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Cryo-electron Microscopy Reconstruction and Stability Studies of the Wild Type and the R432A Variant of Adeno-associated Virus Type 2 Reveal that Capsid Structural Stability Is a Major Factor in Genome Packaging.
J.Virol., 90, 2016
|
|
8P0V
 
 | | Structure of the human Commander complex coiled coils, DENND10 and partial Retriever subcomplex | | Descriptor: | Coiled-coil domain-containing protein 22, Coiled-coil domain-containing protein 93, DENN domain-containing protein 10, ... | | Authors: | Kumpula, E.P, Laulumaa, S, Huiskonen, J.T. | | Deposit date: | 2023-05-10 | | Release date: | 2024-03-20 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Structure and interactions of the endogenous human Commander complex.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4X9N
 
 | | Dithionite reduced L-alpha-Glycerophosphate Oxidase from Mycoplasma pneumoniae with FAD bound | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, L-alpha-glycerophosphate oxidase, NICKEL (II) ION | | Authors: | Elkhal, C.K, Kean, K.M, Parsonage, D, Claiborne, A, Karplus, P.A. | | Deposit date: | 2014-12-11 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structure and proposed mechanism of l-alpha-glycerophosphate oxidase from Mycoplasma pneumoniae.
Febs J., 282, 2015
|
|
