6YRB
 
 | | Crystal structure of the tetramerization domain of the glycoprotein Gn (Andes virus) at pH 7.5 | | Descriptor: | Envelope polyprotein, IODIDE ION, RNA (5'-D(*())-R(P*UP*UP*UP*())-3'), ... | | Authors: | Serris, A, Rey, F.A, Guardado-Calvo, P. | | Deposit date: | 2020-04-20 | | Release date: | 2020-10-14 | | Last modified: | 2020-10-28 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | The Hantavirus Surface Glycoprotein Lattice and Its Fusion Control Mechanism.
Cell, 183, 2020
|
|
8ILA
 
 | | Crystal structure of LmbT from Streptomyces lincolnensis NRRL ISP-5355 in complex with substrates | | Descriptor: | (2~{S})-3-[2-[(2~{S},3~{R},4~{S},5~{R},6~{R})-6-[(1~{R},2~{R})-1-azanyl-2-oxidanyl-propyl]-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanyl-1~{H}-imidazol-5-yl]-2-(trimethyl-$l^{4}-azanyl)propanoic acid, GUANOSINE-5'-DIPHOSPHATE, Glycosyltransferase | | Authors: | Dai, Y, Qiao, H, Xia, M, Fang, P, Liu, W. | | Deposit date: | 2023-03-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Structural Basis of Low-Molecular-Weight Thiol Glycosylation in Lincomycin A Biosynthesis.
Acs Chem.Biol., 18, 2023
|
|
7UN2
 
 | | Crystal structure of a lectin from Canavalia maritima seed (ConM) complexed with Indole-3-butyric acid | | Descriptor: | 3-INDOLEBUTYRIC ACID, CALCIUM ION, CHLORIDE ION, ... | | Authors: | de Sousa, J.P, Bezerra, E.H.S, Sales, M.V, Queiroz, P.P, da Silva, F.M.S, Carvalho, C.P.S, Freire, V.N, Rocha, B.A.M. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of Canavalia maritima lectin complexed with auxins
To Be Published
|
|
4REQ
 
 | | Methylmalonyl-COA Mutase substrate complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, GLYCEROL, ... | | Authors: | Evans, P.R, Mancia, F. | | Deposit date: | 1998-06-17 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational changes on substrate binding to methylmalonyl CoA mutase and new insights into the free radical mechanism.
Structure, 6, 1998
|
|
6ZND
 
 | | [1,2,4]Triazolo[1,5-a]pyrimidine Phosphodiesterase 2 Inhibitors | | Descriptor: | MAGNESIUM ION, ZINC ION, [(3~{S})-3-([1,2,4]triazolo[1,5-a]pyrimidin-7-yl)piperidin-1-yl]-(3,4,5-trimethoxyphenyl)methanone, ... | | Authors: | Tresadern, G, Leonard, P.M. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | [1,2,4]Triazolo[1,5- a ]pyrimidine Phosphodiesterase 2A Inhibitors: Structure and Free-Energy Perturbation-Guided Exploration.
J.Med.Chem., 63, 2020
|
|
6ZNO
 
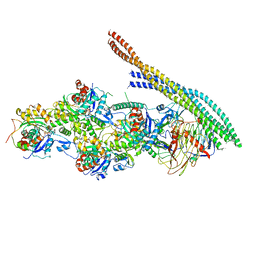 | | The pointed end complex of dynactin with the p150 projection docked | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ARP1 actin related protein 1 homolog A, ... | | Authors: | Lau, C.K, Lacey, S.E, Carter, A.P. | | Deposit date: | 2020-07-06 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Cryo-EM reveals the complex architecture of dynactin's shoulder region and pointed end.
Embo J., 40, 2021
|
|
7SSA
 
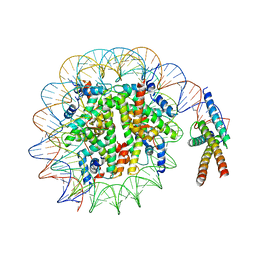 | | Cryo-EM structure of pioneer factor Cbf1 bound to the nucleosome | | Descriptor: | Centromere-binding protein 1, DNA (137-MER), Histone H2A.1, ... | | Authors: | Eek, P, Tan, S. | | Deposit date: | 2021-11-10 | | Release date: | 2023-04-05 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Basic helix-loop-helix pioneer factors interact with the histone octamer to invade nucleosomes and generate nucleosome-depleted regions.
Mol.Cell, 83, 2023
|
|
7SS6
 
 | | Structure of Klebsiella LpxH in complex with JH-LPH-45 | | Descriptor: | 1,2-ETHANEDIOL, 5-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}-N-[5-(hydroxyamino)-5-oxopentyl]-2,3-dihydro-1H-indole-1-carboxamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
7SS7
 
 | | Crystal structure of Klebsiella LpxH in complex with JH-LPH-50 | | Descriptor: | 1,2-ETHANEDIOL, 6-{[(4-{4-[3-chloro-5-(trifluoromethyl)phenyl]piperazine-1-sulfonyl}phenyl)carbamoyl]amino}-N-hydroxyhexanamide, MANGANESE (II) ION, ... | | Authors: | Cho, J, Cochrane, C.S, Zhou, P. | | Deposit date: | 2021-11-09 | | Release date: | 2023-04-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Development of LpxH Inhibitors Chelating the Active Site Dimanganese Metal Cluster of LpxH.
Chemmedchem, 18, 2023
|
|
8VHL
 
 | | Structure of DHODH in Complex with Ligand 17 | | Descriptor: | ACETATE ION, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shaffer, P.L. | | Deposit date: | 2024-01-02 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Discovery of Alternative Binding Poses through Fragment-Based Identification of DHODH Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
6ZPK
 
 | | Crystal structure of the unconventional kinetochore protein Trypanosoma brucei KKT4 BRCT domain | | Descriptor: | GLYCEROL, SULFATE ION, Trypanosoma brucei KKT4 463-645 | | Authors: | Ludzia, P, Lowe, E.D, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
6PEK
 
 | | Structure of Spastin Hexamer (Subunit A-E) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6FHJ
 
 | | Structural dynamics and catalytic properties of a multi-modular xanthanase, native. | | Descriptor: | CALCIUM ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Moroz, O.V, Jensen, P.F, McDonald, S.P, McGregor, N, Blagova, E, Comamala, G, Segura, D.R, Anderson, L, Vasu, S.M, Rao, V.P, Giger, L, Monrad, R.N, Svendsen, A, Nielsen, J.E, Henrissat, B, Davies, G.J, Brumer, H, Rand, K, Wilson, K.S. | | Deposit date: | 2018-01-14 | | Release date: | 2018-08-29 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural Dynamics and Catalytic Properties of a Multimodular Xanthanase
Acs Catalysis, 2018
|
|
6FLF
 
 | |
6WHB
 
 | | MEKK1 TOG domain (548-867) | | Descriptor: | ACETATE ION, GLYCEROL, Mitogen-activated protein kinase kinase kinase 1 | | Authors: | Filipcik, P, Mace, P.D. | | Deposit date: | 2020-04-07 | | Release date: | 2020-08-26 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.90032053 Å) | | Cite: | A cryptic tubulin-binding domain links MEKK1 to curved tubulin protomers.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5F9O
 
 | | Crystal structure of broadly neutralizing VH1-46 germline-derived CD4-binding site-directed antibody CH235.09 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CH235.09 Light chain, ... | | Authors: | Joyce, M.G, Mascola, J.R, Kwong, P.D. | | Deposit date: | 2015-12-10 | | Release date: | 2016-03-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Maturation Pathway from Germline to Broad HIV-1 Neutralizer of a CD4-Mimic Antibody.
Cell, 165, 2016
|
|
6YY4
 
 | | Parallel 17-mer DNA G-quadruplex | | Descriptor: | DNA (5'-D(*GP*GP*GP*TP*GP*GP*GP*AP*AP*GP*GP*GP*TP*GP*GP*GP*A)-3') | | Authors: | Srb, P, Curtis, C, Veverka, V. | | Deposit date: | 2020-05-04 | | Release date: | 2021-01-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Overlapping but distinct: a new model for G-quadruplex biochemical specificity.
Nucleic Acids Res., 49, 2021
|
|
8V87
 
 | | 60S ribosome biogenesis intermediate (Dbp10 post-catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 25S ribosomal RNA, 27S pre-rRNA (guanosine(2922)-2'-O)-methyltransferase, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
5J9T
 
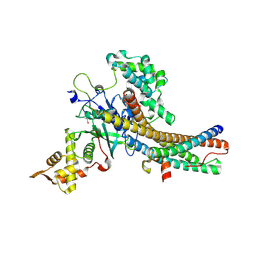 | | Crystal structure of the NuA4 core complex | | Descriptor: | Chromatin modification-related protein EAF6, Chromatin modification-related protein YNG2, Enhancer of polycomb-like protein 1, ... | | Authors: | Chen, Z.C, Xu, P. | | Deposit date: | 2016-04-11 | | Release date: | 2016-10-26 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The NuA4 Core Complex Acetylates Nucleosomal Histone H4 through a Double Recognition Mechanism
Mol.Cell, 63, 2016
|
|
6UHO
 
 | | Crystal Structure of C148 mGFP-cDNA-2 | | Descriptor: | C148 mGFP-cDNA-2, UNKNOWN LIGAND | | Authors: | Winegar, P.W, Hayes, O.G, McMillan, J.R, Figg, C.A, Focia, P.J, Mirkin, C.A. | | Deposit date: | 2019-09-27 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | DNA-Directed Protein Packing within Single Crystals.
Chem, 6, 2020
|
|
6PDW
 
 | | Msp1-substrate complex in closed conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Wang, L, Myasnikov, A, Pan, X, Walter, P. | | Deposit date: | 2019-06-19 | | Release date: | 2020-02-12 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the AAA protein Msp1 reveals mechanism of mislocalized membrane protein extraction.
Elife, 9, 2020
|
|
6PEN
 
 | | Structure of Spastin Hexamer (whole model) in complex with substrate peptide | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, EYEYEYEYEY, ... | | Authors: | Han, H, Schubert, H.L, McCullough, J, Monroe, N, Sundquist, W.I, Hill, C.P. | | Deposit date: | 2019-06-20 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structure of spastin bound to a glutamate-rich peptide implies a hand-over-hand mechanism of substrate translocation.
J.Biol.Chem., 295, 2020
|
|
6WAZ
 
 | | +1 extended HIV-1 reverse transcriptase initiation complex core (pre-translocation state) | | Descriptor: | HIV-1 viral RNA genome fragment, Reverse transcriptase p51 subunit, Reverse transcriptase/ribonuclease H, ... | | Authors: | Larsen, K.P, Jackson, L.N, Kappel, K, Zhang, J, Puglisi, E.V. | | Deposit date: | 2020-03-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Distinct Conformational States Underlie Pausing during Initiation of HIV-1 Reverse Transcription.
J.Mol.Biol., 432, 2020
|
|
7AFR
 
 | | Ribosome maturation factor RimP (apo) | | Descriptor: | Ribosome maturation factor RimP | | Authors: | Schedlbauer, A, Iturrioz, I, Ochoa-Lizarralde, B, Diercks, T, Fucini, P, Connell, S. | | Deposit date: | 2020-09-19 | | Release date: | 2021-07-07 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | A conserved rRNA switch is central to decoding site maturation on the small ribosomal subunit.
Sci Adv, 7, 2021
|
|
