4UT0
 
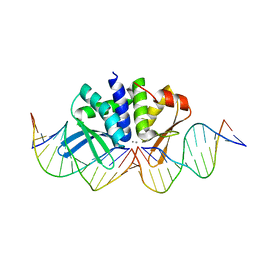 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 10 DAYS INCUBATION IN 5MM MN (STATE 7) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*CP)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*CP)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*AP)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-07-17 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
1N96
 
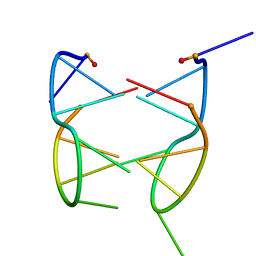 | | DIMERIC SOLUTION STRUCTURE OF THE CYCLIC OCTAMER CD(CGCTCATT) | | Descriptor: | CYCLIC OLIGONUCLEOTIDE D(CGCTCATT) | | Authors: | Escaja, N, Gelpi, J.L, Orozco, M, Rico, M, Pedroso, E, Gonzalez, C. | | Deposit date: | 2002-11-22 | | Release date: | 2003-05-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Four-stranded DNA structure stabilized by a novel G:C:A:T tetrad.
J.Am.Chem.Soc., 125, 2003
|
|
4UN9
 
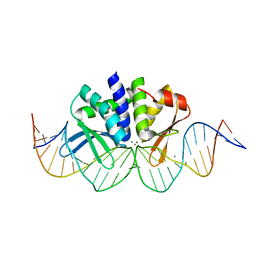 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8H INCUBATION IN 5MM MN (STATE 3) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.734 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNC
 
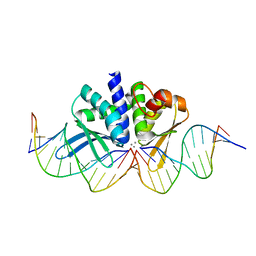 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 8 DAYS INCUBATION IN 5MM MN (STATE 6) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNB
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 6 DAYS INCUBATION IN 5MM MN (STATE 5) | | Descriptor: | 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', 5'-D(*GP*CP*CP*TP*TP*GP*CP*CP*GP*GP*GP*TP*AP*A)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UN7
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA BEFORE INCUBATION IN 5MM MN (STATE 1) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, ZINC ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
8OFC
 
 | | Structure of an i-motif domain with the cytosine analog 1,3-diaza-2-oxophenoxacione (tC) at neutral pH | | Descriptor: | DNA (5'-D(*CP*(YCO)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*(DNR)P*GP*T)-3') | | Authors: | Mir, B, Serrano-Chacon, I, Terrazas, M, Gandioso, A, Garavis, M, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2023-03-15 | | Release date: | 2024-02-07 | | Last modified: | 2024-04-24 | | Method: | SOLUTION NMR | | Cite: | Site-specific incorporation of a fluorescent nucleobase analog enhances i-motif stability and allows monitoring of i-motif folding inside cells.
Nucleic Acids Res., 52, 2024
|
|
5M3D
 
 | | Structural tuning of CD81LEL (space group P31) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, PHOSPHATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-14 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
5M4R
 
 | | Structural tuning of CD81LEL (space group C2) | | Descriptor: | 1,2-ETHANEDIOL, CD81 antigen, SULFATE ION | | Authors: | Cunha, E.S, Sfriso, P, Rojas, A.L, Roversi, P, Hospital, A, Orozco, M, Abrescia, N.G. | | Deposit date: | 2016-10-19 | | Release date: | 2016-12-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism of Structural Tuning of the Hepatitis C Virus Human Cellular Receptor CD81 Large Extracellular Loop.
Structure, 25, 2017
|
|
8QE4
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*CP*(FRG)P*CP*(FRG)P*CP*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QOR
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(CFL)P*(FRG)P*(CFL)P*(FRG)P*(CFL)P*(FRG))-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Friedland, D, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
8QDU
 
 | | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions | | Descriptor: | DNA (5'-D(*(FC)P*GP*(FC)P*GP*(FC)P*G)-3') | | Authors: | El-Khoury, R, Cabrero, C, Movilla, S, Thorpe, J.D, Roman, M, Orozco, M, Gonzalez, C, Damha, M.J. | | Deposit date: | 2023-08-30 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-31 | | Method: | SOLUTION NMR | | Cite: | Formation of left-handed helices by C2'-fluorinated nucleic acids under physiological salt conditions.
Nucleic Acids Res., 52, 2024
|
|
1IEK
 
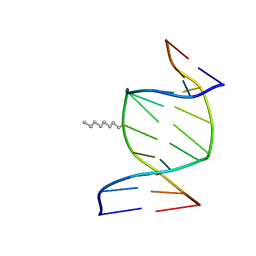 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER S) | | Descriptor: | 5'-D(*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-10 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
1IEY
 
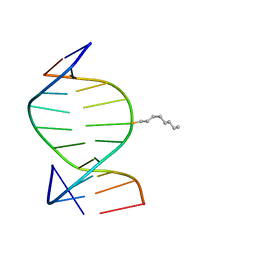 | | SOLUTION STRUCTURE OF THE DNA DUPLEX D(CCACCGGAAC).(GTTCCGGTGG) WITH A CHIRAL ALKYL-PHOSPHONATE MOIETY (DIAESTEREOISOMER R) | | Descriptor: | 5'-D(*GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3', 5'-D(P*CP*CP*AP*CP*CP*(OCT)GP*GP*AP*AP*C)-3', N-OCTANE | | Authors: | Soliva, R, Monaco, V, Gomez-Pinto, I, Meeuwenoord, N.J, van der Marel, G.A, van Boom, J.H, Gonzalez, C, Orozco, M. | | Deposit date: | 2001-04-11 | | Release date: | 2001-07-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a DNA duplex with a chiral alkyl phosphonate moiety.
Nucleic Acids Res., 29, 2001
|
|
8BQY
 
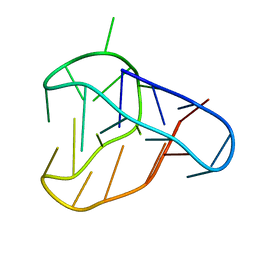 | | An i-motif domain able to undergo pH-dependent conformational transitions (acidic structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*(DNR)P*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*(DNR)P*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-11-22 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
8BV6
 
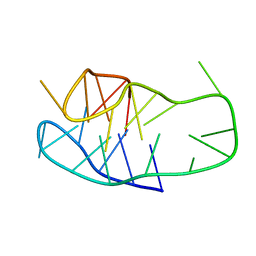 | | An i-motif domain able to undergo pH-dependent conformational transitions (neutral structure) | | Descriptor: | DNA (5'-D(*CP*(DNR)P*GP*TP*TP*CP*(DNR)P*GP*TP*TP*TP*TP*TP*CP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Serrano-Chacon, I, Mir, B, Cupellini, L, Colizzi, F, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2022-12-01 | | Release date: | 2023-02-22 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | pH-Dependent Capping Interactions Induce Large-Scale Structural Transitions in i-Motifs.
J.Am.Chem.Soc., 145, 2023
|
|
1SP6
 
 | | A DNA duplex containing a cholesterol adduct (alpha-anomer) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOL)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-16 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
1SSJ
 
 | | A DNA DUPLEX CONTAINING A CHOLESTEROL ADDUCT (BETA-ANOMER) | | Descriptor: | 5'-D(*CP*CP*AP*CP*(HOB)P*GP*GP*AP*AP*C)-3', 5'-D(GP*TP*TP*CP*CP*GP*GP*TP*GP*G)-3' | | Authors: | Gomez-Pinto, I, Cubero, E, Kalko, S.G, Monaco, V, van der Marel, G, van Boom, J.H, Orozco, M, Gonzalez, C. | | Deposit date: | 2004-03-24 | | Release date: | 2004-06-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Effect of bulky lesions on DNA: Solution structure of a DNA duplex containing a cholesterol adduct.
J.Biol.Chem., 279, 2004
|
|
4UN8
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 1H INCUBATION IN 5MM MN (STATE 2) | | Descriptor: | 25MER, HOMING ENDONUCLEASE I-DMOI, MANGANESE (II) ION | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
4UNA
 
 | | THE CRYSTAL STRUCTURE OF I-DMOI IN COMPLEX WITH ITS TARGET DNA AT 2 DAYS INCUBATION IN 5MM MN (STATE 4) | | Descriptor: | 25MER, 5'-D(*CP*CP*GP*GP*CP*AP*AP*GP*GP*C)-3', 5'-D(*CP*GP*CP*GP*CP*CP*GP*GP*AP*AP*CP*TP*TP*AP*C)-3', ... | | Authors: | Molina, R, Stella, S, Redondo, P, Gomez, H, Marcaida, M.J, Orozco, M, Prieto, J, Montoya, G. | | Deposit date: | 2014-05-26 | | Release date: | 2014-12-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Visualizing Phosphodiester-Bond Hydrolysis by an Endonuclease.
Nat.Struct.Mol.Biol., 22, 2015
|
|
7QDH
 
 | | SARS-CoV-2 S protein S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Fibritin | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7QDG
 
 | | SARS-CoV-2 S protein S:A222V + S:D614G mutant 1-up | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ginex, T, Marco-Marin, C, Wieczor, M, Mata, C.P, Krieger, J, Lopez-Redondo, M.L, Frances-Gomez, C, Ruiz-Rodriguez, P, Melero, R, Sanchez-Sorzano, C.O, Martinez, M, Gougeard, N, Forcada-Nadal, A, Zamora-Caballero, S, Gozalbo-Rovira, R, Sanz-Frasquet, C, Bravo, J, Rubio, V, Marina, A, Geller, R, Comas, I, Gil, C, Coscolla, M, Orozco, M, LLacer, J.L, Carazo, J.M. | | Deposit date: | 2021-11-27 | | Release date: | 2022-05-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The structural role of SARS-CoV-2 genetic background in the emergence and success of spike mutations: The case of the spike A222V mutation.
Plos Pathog., 18, 2022
|
|
7OHE
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*TP*CP*GP*AP*CP*GP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-10 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHM
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*AP*(DH)P*GP*TP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
7OHJ
 
 | | A self-complementary DNA dodecamer duplex contaning 5-hydroxymethylcitosine | | Descriptor: | DNA (5'-D(*CP*GP*AP*CP*GP*TP*CP*G)-3') | | Authors: | Battistini, F, Dans, P.D, Terrazas, M, Castellazzi, C.L, Portella, G, Labrador, M, Villegas, N, Brun-Heath, I, Gonzalez, C, Orozco, M. | | Deposit date: | 2021-05-11 | | Release date: | 2021-11-03 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The Impact of the HydroxyMethylCytosine epigenetic signature on DNA structure and function.
Plos Comput.Biol., 17, 2021
|
|
