3I1F
 
 | | Gamma-subunit of the translation initiation factor 2 from S. solfataricus in complex with Gpp(CH2)p | | Descriptor: | PHOSPHATE ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, Translation initiation factor 2 subunit gamma | | Authors: | Stolboushkina, E.A, Nikonov, S.V, Nikulin, A.D, Blaesi, U, Garber, M.B, Nikonov, O.S. | | Deposit date: | 2009-06-26 | | Release date: | 2010-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 5-prime-triphosphate end of mRNA recognizes the second GTP-binding site on the gamma-subunit of the archaeal translation initiation factor 2
To be Published
|
|
4M0L
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus complexed with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Arkhipova, V.I, Gabdulkhakov, A.G, Nikulin, A.D, Lazopulo, A.M, Lazopulo, S.M, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2013-08-01 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational transitions in the gamma subunit of the archaeal translation initiation factor 2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
5B4Z
 
 | | Crystal structure of heme binding protein HmuT H141A mutant | | Descriptor: | ABC-type transporter, periplasmic component, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2016-04-20 | | Release date: | 2017-03-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structural Characterization of Heme Environmental Mutants of CgHmuT that Shuttles Heme Molecules to Heme Transporters
Int J Mol Sci, 17, 2016
|
|
4NBS
 
 | | The structure of aIF2gamma subunit H20F from archaeon Sulfolobus solfataricus complexed with GDPCP | | Descriptor: | FORMIC ACID, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID GUANYLATE ESTER, ... | | Authors: | Kravchenko, O.V, Nikonov, O.S, Arhipova, V.I, Stolboushkina, E.A, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2013-10-23 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.305 Å) | | Cite: | The structure of aIF2gamma subunit H20F from archaeon Sulfolobus solfataricus complexed with GDPCP
To be Published
|
|
4M2L
 
 | | Gamma subunit of the translation initiation factor 2 from Sulfolobus solfataricus in nucleotide-free form | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, SULFATE ION, ... | | Authors: | Nikonov, O.S, Stolboushkina, E.A, Arkhipova, V.I, Gabdulkhakov, A.G, Nikulin, A.D, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2013-08-05 | | Release date: | 2014-03-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.149 Å) | | Cite: | Conformational transitions in the gamma subunit of the archaeal translation initiation factor 2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
7DVS
 
 | | Crystal structure of Apo (heme-free) PefR | | Descriptor: | MarR family transcriptional regulator | | Authors: | Muraki, N, Aono, S. | | Deposit date: | 2021-01-15 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Heme controls the structural rearrangement of its sensor protein mediating the hemolytic bacterial survival.
Commun Biol, 4, 2021
|
|
4X9C
 
 | | 1.4A crystal structure of Hfq from Methanococcus jannaschii | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Nikulin, A.D, Tishchenko, S.V, Nikonova, S.V, Murina, V.N, Mihailina, A.O, Lekontseva, N.V. | | Deposit date: | 2014-12-11 | | Release date: | 2014-12-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Characterization of RNA-binding properties of the archaeal Hfq-like protein from Methanococcus jannaschii.
J. Biomol. Struct. Dyn., 35, 2017
|
|
1RIS
 
 | | CRYSTAL STRUCTURE OF THE RIBOSOMAL PROTEIN S6 FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Lindahl, M, Svensson, L.A, Liljas, A, Sedelnikova, S.E, Eliseikina, I.A, Fomenkova, N.P, Nevskaya, N, Nikonov, S.V, Garber, M.B, Muranova, T.A, Rykonova, A.I, Amons, R. | | Deposit date: | 1994-05-31 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ribosomal protein S6 from Thermus thermophilus.
EMBO J., 13, 1994
|
|
7DIH
 
 | |
1FEU
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN TL5, ONE OF THE CTC FAMILY PROTEINS, COMPLEXED WITH A FRAGMENT OF 5S RRNA. | | Descriptor: | 19 NT FRAGMENT OF 5S RRNA, 21 NT FRAGMENT OF 5S RRNA, 50S RIBOSOMAL PROTEIN L25, ... | | Authors: | Fedorov, R.V, Meshcheryakov, V.A, Gongadze, G.M, Fomenkova, N.P, Nevskaya, N.A, Selmer, M, Laurberg, M, Kristensen, O, Al-Karadaghi, S, Liljas, A, Garber, M.B, Nikonov, S.V. | | Deposit date: | 2000-07-23 | | Release date: | 2001-06-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of ribosomal protein TL5 complexed with RNA provides new insights into the CTC family of stress proteins.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
1ZHO
 
 | | The structure of a ribosomal protein L1 in complex with mRNA | | Descriptor: | 50S ribosomal protein L1, POTASSIUM ION, mRNA | | Authors: | Nevskaya, N, Tishchenko, S, Volchkov, S, Kljashtorny, V, Nikonova, E, Nikonov, O, Nikulin, A, Kohrer, C, Piendl, W, Zimmermann, R, Stockley, P, Garber, M, Nikonov, S. | | Deposit date: | 2005-04-26 | | Release date: | 2006-05-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New insights into the interaction of ribosomal protein L1 with RNA.
J.Mol.Biol., 355, 2006
|
|
2HW8
 
 | | Structure of ribosomal protein L1-mRNA complex at 2.1 resolution. | | Descriptor: | 1,4-BUTANEDIOL, 36-MER, 50S ribosomal protein L1, ... | | Authors: | Tishchenko, S, Nikonova, E, Nikulin, A, Nevskaya, N, Volchkov, S, Piendl, W, Garber, M, Nikonov, S. | | Deposit date: | 2006-08-01 | | Release date: | 2006-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the ribosomal protein L1-mRNA complex at 2.1 A resolution: common features of crystal packing of L1-RNA complexes.
ACTA CRYSTALLOGR.,SECT.D, 62, 2006
|
|
7FEH
 
 | | Crystal structure of human DDR1 in complex with CH5541127 | | Descriptor: | Epithelial discoidin domain-containing receptor 1, N-[(5-chloranyl-2-ethylsulfonyl-phenyl)methyl]-3-piperazin-1-yl-5-(trifluoromethyloxy)benzamide, NITRATE ION | | Authors: | Fukami, T.A, Kadono, S, Matsuura, T. | | Deposit date: | 2021-07-20 | | Release date: | 2022-07-27 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Novel potent and highly selective DDR1 inhibitors from integrated lead finding
Medicinal Chemistry Research, 32, 2023
|
|
4QG3
 
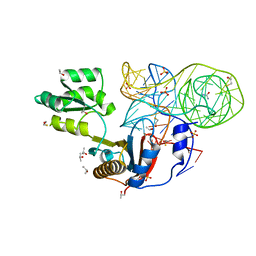 | | Crystal structure of mutant ribosomal protein G219V TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 50S ribosomal protein L1, BETA-MERCAPTOETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
1F7Y
 
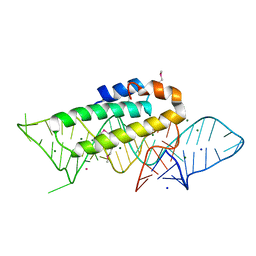 | | THE CRYSTAL STRUCTURE OF TWO UUCG LOOPS HIGHLIGHTS THE ROLE PLAYED BY 2'-HYDROXYL GROUPS IN ITS UNUSUAL STABILITY | | Descriptor: | 16S RIBOSOMAL RNA FRAGMENT, 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, ... | | Authors: | Ennifar, E, Nikouline, A, Serganov, A, Tishchenko, S, Nevskaya, N, Garber, M, Ehresmann, B, Ehresmann, C, Nikonov, S, Dumas, P. | | Deposit date: | 2000-06-28 | | Release date: | 2000-11-22 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The crystal structure of UUCG tetraloop.
J.Mol.Biol., 304, 2000
|
|
4REO
 
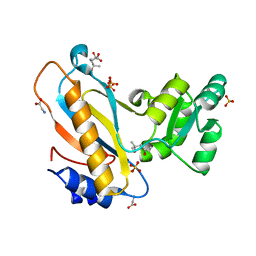 | | Mutant ribosomal protein l1 from thermus thermophilus with threonine 217 replaced by valine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 50S ribosomal protein L1, GLYCINE, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Tishchenko, S.V, Nikonov, S.V. | | Deposit date: | 2014-09-23 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
7F30
 
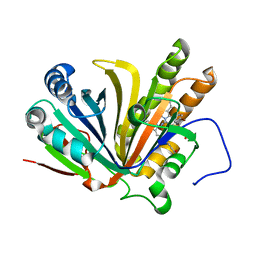 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Y
 
 | | Crystal structure of OxdB E85A mutant (form I) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
7F2Z
 
 | | Crystal structure of OxdB E85A mutant (form II) | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
4QVI
 
 | | Crystal structure of mutant ribosomal protein M218L TthL1 in complex with 80nt 23S RNA from Thermus thermophilus | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 50S ribosomal protein L1, ACETATE ION, ... | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, NIkonov, S.V. | | Deposit date: | 2014-07-15 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
4QGB
 
 | | Crystal structure of mutant ribosomal protein G219V TthL1 | | Descriptor: | 50S ribosomal protein L1, ACETATE ION, CHLORIDE ION | | Authors: | Gabdulkhakov, A.G, Nevskaya, N.A, Nikonov, S.V. | | Deposit date: | 2014-05-22 | | Release date: | 2015-02-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Protein-RNA affinity of ribosomal protein L1 mutants does not correlate with the number of intermolecular interactions.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
