8HM2
 
 | |
8HM1
 
 | |
3UZC
 
 | | Thermostabilised Adenosine A2A receptor in complex with 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol | | Descriptor: | 4-(3-amino-5-phenyl-1,2,4-triazin-6-yl)-2-chlorophenol, Adenosine A2A Receptor | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.341 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
3UZA
 
 | | Thermostabilised Adenosine A2A receptor in complex with 6-(2,6-Dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine | | Descriptor: | 6-(2,6-dimethylpyridin-4-yl)-5-phenyl-1,2,4-triazin-3-amine, Adenosine receptor A2a | | Authors: | Congreve, M, Andrews, S.P, Dore, A.S, Hollenstein, K, Hurrell, E, Langmead, C.J, Mason, J.S, Ng, I.W, Tehan, B, Zhukov, A, Weir, M, Marshall, F.H. | | Deposit date: | 2011-12-07 | | Release date: | 2012-03-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.273 Å) | | Cite: | Discovery of 1,2,4-Triazine Derivatives as Adenosine A(2A) Antagonists using Structure Based Drug Design
J.Med.Chem., 55, 2012
|
|
1MKA
 
 | |
2N8D
 
 | |
4LDK
 
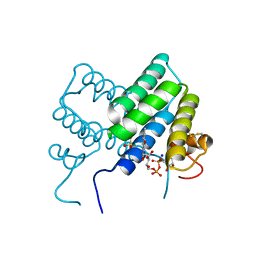 | |
3UN9
 
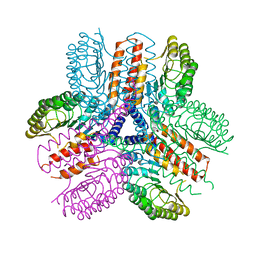 | | Crystal structure of an immune receptor | | Descriptor: | NLR family member X1, PLATINUM (II) ION | | Authors: | Hong, M, Yoon, S.I, Wilson, I.A. | | Deposit date: | 2011-11-15 | | Release date: | 2012-03-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure and Functional Characterization of the RNA-Binding Element of the NLRX1 Innate Immune Modulator.
Immunity, 36, 2012
|
|
1MKB
 
 | |
3U5S
 
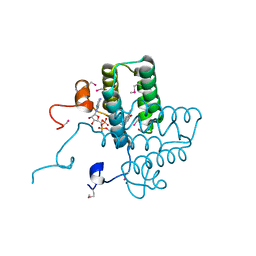 | |
4M5Z
 
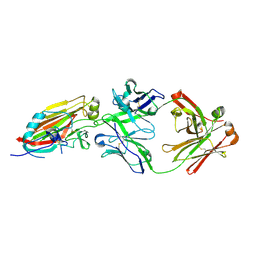 | | Crystal structure of broadly neutralizing antibody 5J8 bound to 2009 pandemic influenza hemagglutinin, HA1 subunit | | Descriptor: | Fab 5J8 heavy chain, Fab 5J8 light chain, Hemagglutinin HA1 chain | | Authors: | Hong, M, Lee, P.S, Wilson, I.A. | | Deposit date: | 2013-08-08 | | Release date: | 2013-09-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Antibody Recognition of the Pandemic H1N1 Influenza Virus Hemagglutinin Receptor Binding Site.
J.Virol., 87, 2013
|
|
3TK0
 
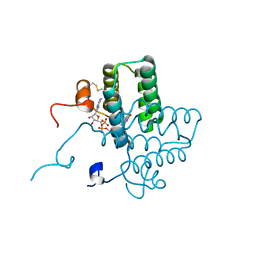 | | mutation of sfALR | | Descriptor: | FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Dong, M, Bahnson, B.J. | | Deposit date: | 2011-08-25 | | Release date: | 2012-08-29 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.611 Å) | | Cite: | sfALR mutation
To be Published
|
|
2G38
 
 | | A PE/PPE Protein Complex from Mycobacterium tuberculosis | | Descriptor: | MANGANESE (II) ION, PE FAMILY PROTEIN, PPE FAMILY PROTEIN | | Authors: | Strong, M, Sawaya, M.R, Eisenberg, D, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2006-02-17 | | Release date: | 2006-03-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Toward the structural genomics of complexes: Crystal structure of a PE/PPE protein complex from Mycobacterium tuberculosis.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2KAD
 
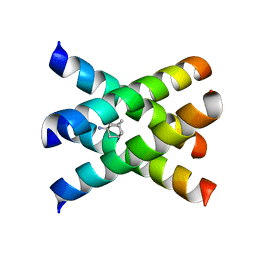 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
2JJC
 
 | | Hsp90 alpha ATPase domain with bound small molecule fragment | | Descriptor: | DIMETHYL SULFOXIDE, HEAT SHOCK PROTEIN HSP 90-ALPHA, PYRIMIDIN-2-AMINE | | Authors: | Congreve, M, Chessari, G, Tisi, D, Woodhead, A.J. | | Deposit date: | 2008-03-31 | | Release date: | 2008-07-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Recent Developments in Fragment-Based Drug Discovery.
J.Med.Chem., 51, 2008
|
|
6VED
 
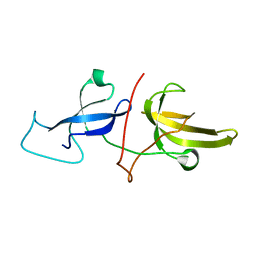 | | Solution structure of the TTD and linker region of UHRF1 | | Descriptor: | E3 ubiquitin-protein ligase UHRF1 | | Authors: | Lemak, A, Houliston, S, Duan, S, Ong, M.S, Arrowsmith, C.H. | | Deposit date: | 2019-12-31 | | Release date: | 2020-06-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Alternative splicing and allosteric regulation modulate the chromatin binding of UHRF1.
Nucleic Acids Res., 48, 2020
|
|
5WYR
 
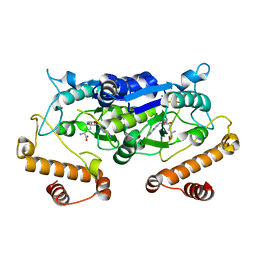 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
5WYQ
 
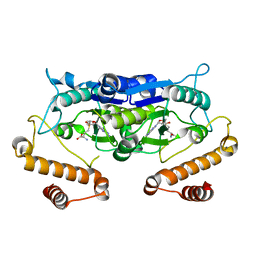 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
4PYZ
 
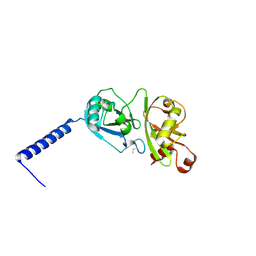 | | Crystal structure of the first two Ubl domains of Deubiquitylase USP7 | | Descriptor: | UNKNOWN ATOM OR ION, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Walker, J.R, Dong, A, Ong, M.S, Dhe-Paganon, S, Kania, J, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-03-28 | | Release date: | 2014-04-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Crystal structure of the first two Ubl domains of Deubiquitylase USP7
to be published
|
|
4TVR
 
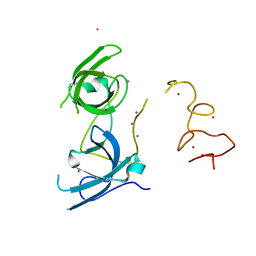 | | Tandem Tudor and PHD domains of UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Ong, M, Duan, S, Li, Y, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Tandem Tudor and PHD domains of UHRF2
To be published
|
|
6TP3
 
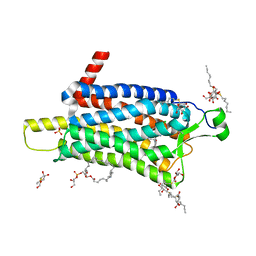 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TO7
 
 | | Crystal structure of the Orexin-1 receptor in complex with suvorexant at 2.29 A resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
1ZY6
 
 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
6TPJ
 
 | | Crystal structure of the Orexin-2 receptor in complex with suvorexant at 2.76 A resolution | | Descriptor: | AMMONIUM ION, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQ7
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-334867 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-(2-methyl-1,3-benzoxazol-6-yl)-3-(1,5-naphthyridin-4-yl)urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6636 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
