7F1K
 
 | | Designed enzyme RA61 M48K/I72D mutant: form IV | | Descriptor: | Engineered Retroaldolase | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
7F1L
 
 | | Designed enzyme RA61 M48K/I72D mutant: form V | | Descriptor: | CHLORIDE ION, Engineered Retroaldolase, IMIDAZOLE | | Authors: | Fujioka, T, Oka, M, Numoto, N, Ito, N, Oda, M, Tanaka, F. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Varying the Directionality of Protein Catalysts for Aldol and Retro-Aldol Reactions.
Chembiochem, 23, 2022
|
|
1X98
 
 | | Crystal structure of Aldose Reductase complexed with 2S4R (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2S,4R)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, CITRIC ACID, ... | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X97
 
 | | Crystal structure of Aldose Reductase complexed with 2R4S (Stereoisomer of Fidarestat, 2S4S) | | Descriptor: | (2R,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X96
 
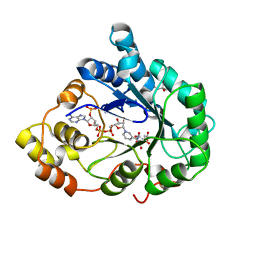 | | Crystal structure of Aldose Reductase with citrates bound in the active site | | Descriptor: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | Authors: | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-09-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
3WQH
 
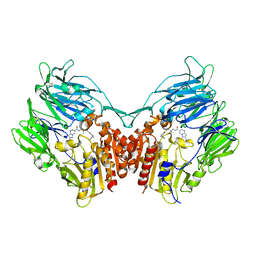 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | Authors: | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | Deposit date: | 2014-01-27 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
4H8N
 
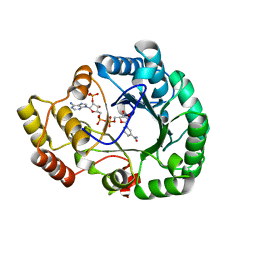 | | Crystal structure of conjugated polyketone reductase C2 from candida parapsilosis complexed with NADPH | | Descriptor: | Conjugated polyketone reductase C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | Deposit date: | 2012-09-23 | | Release date: | 2013-08-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | Descriptor: | Non-structural protein 11 | | Authors: | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-11-16 | | Release date: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.898 Å) | | Cite: | Crystal structure of MERS-CoV main protease
To Be Published
|
|
4WH2
 
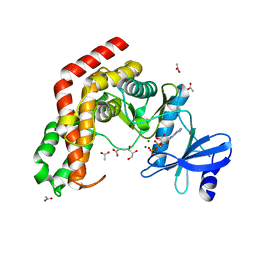 | | N-acetylhexosamine 1-kinase in complex with ADP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.847 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
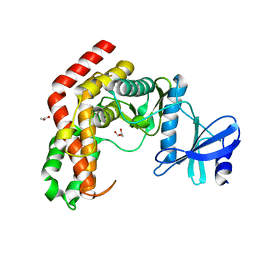 | | N-Acetylhexosamine 1-kinase (ligand free) | | Descriptor: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-11 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
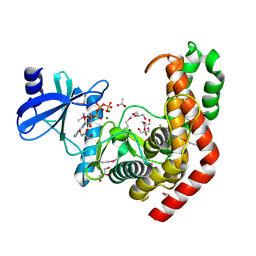 | | N-acetylhexosamine 1-kinase in complex with ATP | | Descriptor: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4TMC
 
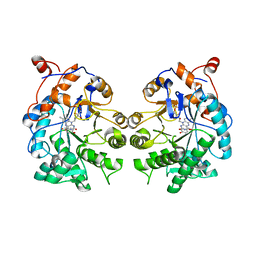 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 COMPLEXED with P-HYDROXYBENZALDEHYDE | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme, P-HYDROXYBENZALDEHYDE | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4TMB
 
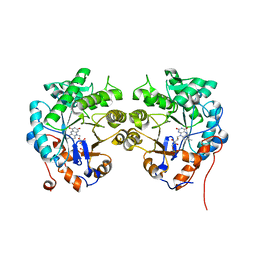 | | CRYSTAL STRUCTURE of OLD YELLOW ENZYME from CANDIDA MACEDONIENSIS AKU4588 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old yellow enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2014-05-31 | | Release date: | 2015-02-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Engineered Old Yellow Enzyme that Enables Efficient Synthesis of (4R,6R)-Actinol in a One-Pot Reduction System
Chembiochem, 16, 2015
|
|
4UNI
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-05-28 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOQ
 
 | | Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | Descriptor: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-09 | | Release date: | 2014-10-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOZ
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose | | Descriptor: | BETA-GALACTOSIDASE, TRIETHYLENE GLYCOL, ZINC ION, ... | | Authors: | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | Deposit date: | 2014-06-11 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
1UC4
 
 | | Structure of diol dehydratase complexed with (S)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.BIOL.CHEM., 278, 2003
|
|
1V7O
 
 | |
1UC5
 
 | | Structure of diol dehydratase complexed with (R)-1,2-propanediol | | Descriptor: | AMMONIUM ION, CYANOCOBALAMIN, POTASSIUM ION, ... | | Authors: | Shibata, N, Nakanishi, Y, Fukuoka, M, Yamanishi, M, Yasuoka, N, Toraya, T. | | Deposit date: | 2003-04-08 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural rationalization for the lack of stereospecificity in coenzyme B12-dependent diol dehydratase
J.Biol.Chem., 278, 2003
|
|
8WWT
 
 | |
8WUP
 
 | |
8WW5
 
 | |
8WX6
 
 | |
6YBT
 
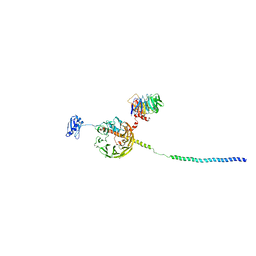 | | Structure of a human 48S translational initiation complex - eIF3bgi | | Descriptor: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit I | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
6YBV
 
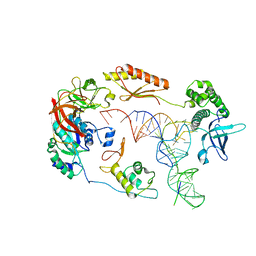 | | Structure of a human 48S translational initiation complex - eIF2-TC | | Descriptor: | Eukaryotic translation initiation factor 2 subunit 1, Eukaryotic translation initiation factor 2 subunit 2, Eukaryotic translation initiation factor 2 subunit 3, ... | | Authors: | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | Deposit date: | 2020-03-17 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
