1PWM
 
 | | Crystal structure of human Aldose Reductase complexed with NADP and Fidarestat | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | El-Kabbani, O, Darmanin, C, Schneider, T.R, Hazemann, I, Ruiz, F, Oka, M, Joachimiak, A, Schulze-Briese, C, Tomizaki, T, Mitschler, A, Podjarny, A. | | 登録日 | 2003-07-02 | | 公開日 | 2004-02-24 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | Ultrahigh resolution drug design. II. Atomic resolution structures of human aldose reductase holoenzyme complexed with Fidarestat and Minalrestat: implications for the binding of cyclic imide inhibitors
PROTEINS, 55, 2004
|
|
2AGT
 
 | | Aldose Reductase Mutant Leu 300 Pro complexed with Fidarestat | | 分子名称: | (2S,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose reductase, CHLORIDE ION, ... | | 著者 | Petrova, T, Steuber, H, Hazemann, I, Cousido-Siah, A, Mitschler, A, Chung, R, Oka, M, Klebe, G, El-Kabbani, O, Joachimiak, A, Podjarny, A. | | 登録日 | 2005-07-27 | | 公開日 | 2005-09-20 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1 Å) | | 主引用文献 | Factorizing Selectivity Determinants of Inhibitor Binding toward Aldose and Aldehyde Reductases: Structural and Thermodynamic Properties of the Aldose Reductase Mutant Leu300Pro-Fidarestat Complex
J.Med.Chem., 48, 2005
|
|
1X98
 
 | | Crystal structure of Aldose Reductase complexed with 2S4R (Stereoisomer of Fidarestat, 2S4S) | | 分子名称: | (2S,4R)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, CITRIC ACID, ... | | 著者 | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | 登録日 | 2004-08-19 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X97
 
 | | Crystal structure of Aldose Reductase complexed with 2R4S (Stereoisomer of Fidarestat, 2S4S) | | 分子名称: | (2R,4S)-2-AMINOFORMYL-6-FLUORO-SPIRO[CHROMAN-4,4'-IMIDAZOLIDINE]-2',5'-DIONE, Aldose Reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | 登録日 | 2004-08-19 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
1X96
 
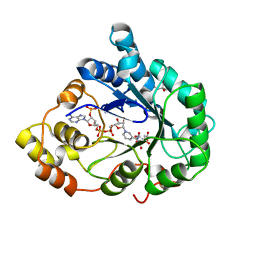 | | Crystal structure of Aldose Reductase with citrates bound in the active site | | 分子名称: | CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, aldose reductase | | 著者 | El-Kabbani, O, Darmanin, C, Oka, M, Schulze-Briese, C, Tomizaki, T, Hazemann, I, Mitschler, A, Podjarny, A. | | 登録日 | 2004-08-19 | | 公開日 | 2004-09-07 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | High-Resolution Structures of Human Aldose Reductase Holoenzyme in Complex with Stereoisomers of the Potent Inhibitor Fidarestat: Stereospecific Interaction between the Enzyme and a Cyclic Imide Type Inhibitor
J.Med.Chem., 47, 2004
|
|
3WQH
 
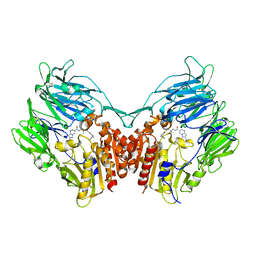 | | Crystal Structure of human DPP-IV in complex with Anagliptin | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Dipeptidyl peptidase 4, N-[2-({2-[(2S)-2-cyanopyrrolidin-1-yl]-2-oxoethyl}amino)-2-methylpropyl]-2-methylpyrazolo[1,5-a]pyrimidine-6-carboxamide | | 著者 | Watanabe, Y.S, Okada, S, Motoyama, T, Takahashi, R, Adachi, H, Oka, M. | | 登録日 | 2014-01-27 | | 公開日 | 2015-07-15 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Anagliptin, a potent dipeptidyl peptidase IV inhibitor: its single-crystal structure and enzyme interactions.
J Enzyme Inhib Med Chem, 30, 2015
|
|
4YPJ
 
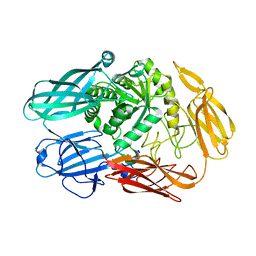 | | X-ray Structure of The Mutant of Glycoside Hydrolase | | 分子名称: | Beta galactosidase | | 著者 | Ishikawa, K, Kataoka, M, Yanamoto, T, Nakabayashi, M, Watanabe, M. | | 登録日 | 2015-03-13 | | 公開日 | 2015-04-29 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of beta-galactosidase from Bacillus circulans ATCC 31382 (BgaD) and the construction of the thermophilic mutants.
Febs J., 282, 2015
|
|
6KU3
 
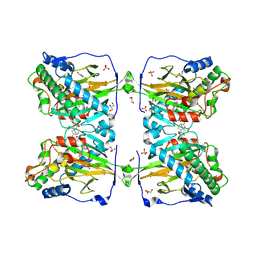 | | Crystal structure of gibberellin 2-oxidase3 (GA2ox3)in rice | | 分子名称: | 2-OXOGLUTARIC ACID, GIBBERELLIN A4, GLYCEROL, ... | | 著者 | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | 登録日 | 2019-08-30 | | 公開日 | 2020-05-13 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
6KUN
 
 | | Crystal structure of dioxygenase for auxin oxidation (DAO) in rice | | 分子名称: | 1H-INDOL-3-YLACETIC ACID, 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent dioxygenase DAO, ... | | 著者 | Takehara, S, Mikami, B, Sakuraba, S, Matsuoka, M, Ueguchi-Tanaka, M. | | 登録日 | 2019-09-02 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.002 Å) | | 主引用文献 | A common allosteric mechanism regulates homeostatic inactivation of auxin and gibberellin.
Nat Commun, 11, 2020
|
|
4UNI
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 in complex with galactose | | 分子名称: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | 登録日 | 2014-05-28 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOQ
 
 | | Nucleophile mutant (E324A) of beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 | | 分子名称: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, TETRAETHYLENE GLYCOL, ... | | 著者 | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | 登録日 | 2014-06-09 | | 公開日 | 2014-10-22 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
4UOZ
 
 | | beta-(1,6)-galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 nucleophile mutant E324A in complex with galactose | | 分子名称: | BETA-GALACTOSIDASE, TRIETHYLENE GLYCOL, ZINC ION, ... | | 著者 | Viborg, A.H, Fredslund, F, Katayama, T, Nielsen, S.K, Svensson, B, Kitaoka, M, Lo Leggio, L, Abou Hachem, M. | | 登録日 | 2014-06-11 | | 公開日 | 2014-10-15 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A beta 1-6/ beta 1-3 galactosidase from Bifidobacterium animalis subsp. lactis Bl-04 gives insight into sub-specificities of beta-galactoside catabolism within Bifidobacterium.
Mol. Microbiol., 2014
|
|
2KSF
 
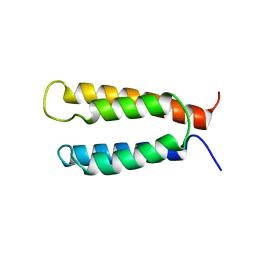 | | Backbone structure of the membrane domain of E. coli histidine kinase receptor KdpD, Center for Structures of Membrane Proteins (CSMP) target 4312C | | 分子名称: | Sensor protein kdpD | | 著者 | Maslennikov, I, Klammt, C, Kefala, G, Okamura, M, Esquivies, L, Kwiatkowski, W, Choe, S, Center for Structures of Membrane Proteins (CSMP) | | 登録日 | 2010-01-03 | | 公開日 | 2010-03-02 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Membrane domain structures of three classes of histidine kinase receptors by cell-free expression and rapid NMR analysis.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
4H8N
 
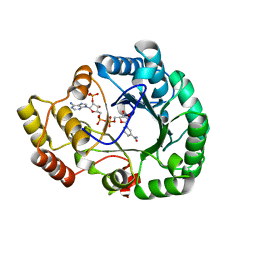 | | Crystal structure of conjugated polyketone reductase C2 from candida parapsilosis complexed with NADPH | | 分子名称: | Conjugated polyketone reductase C2, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Qin, H.-M, Yamamura, A, Miyakawa, T, Maruoka, S, Ohtsuka, J, Nagata, K, Kataoka, M, Shimizu, S, Tanokura, M. | | 登録日 | 2012-09-23 | | 公開日 | 2013-08-07 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure of conjugated polyketone reductase from Candida parapsilosis IFO 0708 reveals conformational changes for substrate recognition upon NADPH binding
Appl.Microbiol.Biotechnol., 98, 2014
|
|
4WH2
 
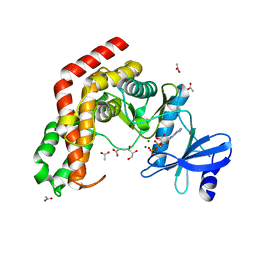 | | N-acetylhexosamine 1-kinase in complex with ADP | | 分子名称: | ACETIC ACID, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | 登録日 | 2014-09-19 | | 公開日 | 2015-02-18 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.847 Å) | | 主引用文献 | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH1
 
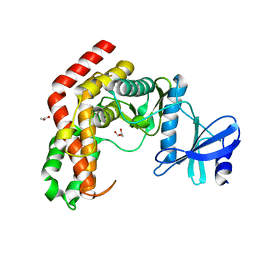 | | N-Acetylhexosamine 1-kinase (ligand free) | | 分子名称: | ACETIC ACID, GLYCEROL, N-acetylhexosamine 1-kinase | | 著者 | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | 登録日 | 2014-09-19 | | 公開日 | 2015-02-11 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
4WH3
 
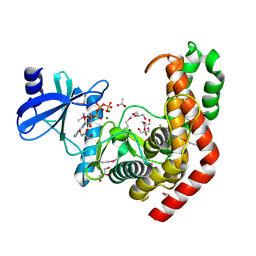 | | N-acetylhexosamine 1-kinase in complex with ATP | | 分子名称: | ACETIC ACID, ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | 著者 | Sato, M, Arakawa, T, Nam, Y.W, Nishimoto, M, Kitaoka, M, Fushinobu, S. | | 登録日 | 2014-09-19 | | 公開日 | 2015-02-18 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Open-close structural change upon ligand binding and two magnesium ions required for the catalysis of N-acetylhexosamine 1-kinase
Biochim.Biophys.Acta, 1854, 2015
|
|
8R5J
 
 | | Crystal structure of MERS-CoV main protease | | 分子名称: | Non-structural protein 11 | | 著者 | Balcomb, B.H, Fairhead, M, Koekemoer, L, Lithgo, R.M, Aschenbrenner, J.C, Chandran, A.V, Godoy, A.S, Lukacik, P, Marples, P.G, Mazzorana, M, Ni, X, Strain-Damerell, C, Thompson, W, Tomlinson, C.W.E, Wild, C, Winokan, M, Fearon, D, Walsh, M.A, von Delft, F. | | 登録日 | 2023-11-16 | | 公開日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (1.898 Å) | | 主引用文献 | Crystal structure of MERS-CoV main protease
To Be Published
|
|
8I09
 
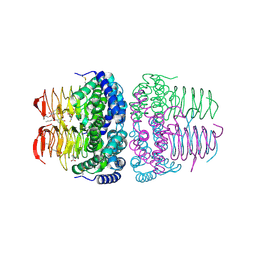 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with butyl gallate | | 分子名称: | CYSTEINE, PHOSPHATE ION, Serine acetyltransferase, ... | | 著者 | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | 登録日 | 2023-01-10 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I06
 
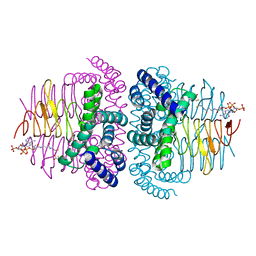 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with CoA | | 分子名称: | COENZYME A, CYSTEINE, Serine acetyltransferase | | 著者 | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | 登録日 | 2023-01-10 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
8I04
 
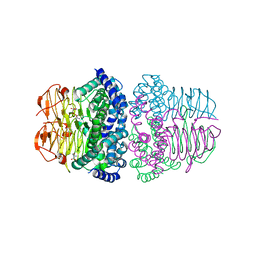 | | Crystal structure of serine acetyltransferase from Salmonella typhimurium complexed with serine | | 分子名称: | PHOSPHATE ION, SERINE, Serine acetyltransferase | | 著者 | Toyomoto, T, Ono, K, Shiba, T, Momitani, K, Zhang, T, Tsutsuki, H, Ishikawa, T, Hoso, K, Hamada, K, Rahman, A, Zhong, H, Akaike, T, Yamamoto, K, Matsuoka, M, Hanaoka, K, Niidome, T, Sawa, T. | | 登録日 | 2023-01-10 | | 公開日 | 2023-11-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Alkyl gallates inhibit serine O -acetyltransferase in bacteria and enhance susceptibility of drug-resistant Gram-negative bacteria to antibiotics.
Front Microbiol, 14, 2023
|
|
1C1B
 
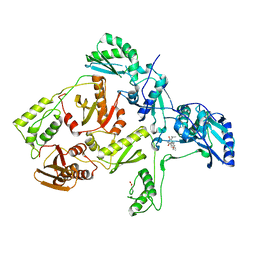 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH GCA-186 | | 分子名称: | 6-(3',5'-DIMETHYLBENZYL)-1-ETHOXYMETHYL-5-ISOPROPYLURACIL, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Hopkins, A.L, Ren, J, Tanaka, H, Baba, B, Okamato, M, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-21 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1C1C
 
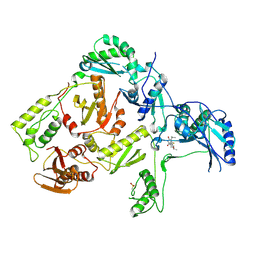 | | CRYSTAL STRUCTURE OF HIV-1 REVERSE TRANSCRIPTASE IN COMPLEX WITH TNK-6123 | | 分子名称: | 6-(cyclohexylsulfanyl)-1-(ethoxymethyl)-5-(1-methylethyl)pyrimidine-2,4(1H,3H)-dione, HIV-1 REVERSE TRANSCRIPTASE (A-CHAIN), HIV-1 REVERSE TRANSCRIPTASE (B-CHAIN) | | 著者 | Hopkins, A.L, Ren, J, Tanaka, H, Baba, M, Okamato, M, Stuart, D.I, Stammers, D.K. | | 登録日 | 1999-07-21 | | 公開日 | 2000-07-21 | | 最終更新日 | 2014-11-12 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Design of MKC-442 (emivirine) analogues with improved activity against drug-resistant HIV mutants.
J.Med.Chem., 42, 1999
|
|
1D8L
 
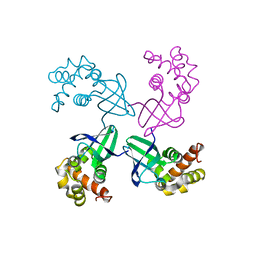 | | E. COLI HOLLIDAY JUNCTION BINDING PROTEIN RUVA NH2 REGION LACKING DOMAIN III | | 分子名称: | PROTEIN (HOLLIDAY JUNCTION DNA HELICASE RUVA) | | 著者 | Nishino, T, Iwasaki, H, Kataoka, M, Ariyoshi, M, Fujita, T, Shinagawa, H, Morikawa, K. | | 登録日 | 1999-10-25 | | 公開日 | 2000-05-03 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Modulation of RuvB function by the mobile domain III of the Holliday junction recognition protein RuvA.
J.Mol.Biol., 298, 2000
|
|
6YBT
 
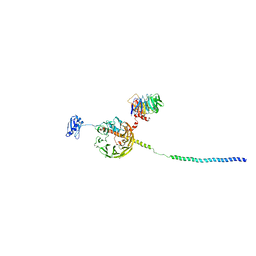 | | Structure of a human 48S translational initiation complex - eIF3bgi | | 分子名称: | Eukaryotic translation initiation factor 3 subunit A, Eukaryotic translation initiation factor 3 subunit B, Eukaryotic translation initiation factor 3 subunit I | | 著者 | Brito Querido, J, Sokabe, M, Kraatz, S, Gordiyenko, Y, Skehel, M, Fraser, C, Ramakrishnan, V. | | 登録日 | 2020-03-17 | | 公開日 | 2020-09-16 | | 最終更新日 | 2024-05-22 | | 実験手法 | ELECTRON MICROSCOPY (6 Å) | | 主引用文献 | Structure of a human 48Stranslational initiation complex.
Science, 369, 2020
|
|
