6LFX
 
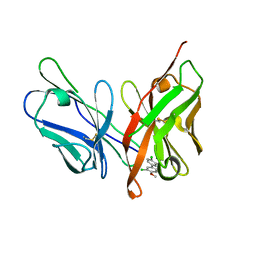 | |
2LQH
 
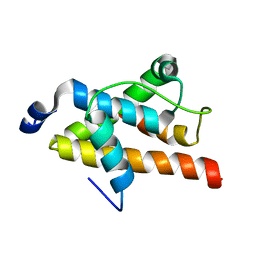 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2b3l conformation) | | 分子名称: | CREB-binding protein, Forkhead box O3 | | 著者 | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | 登録日 | 2012-03-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
2D2P
 
 | | The solution structure of micelle-bound peptide | | 分子名称: | Pituitary adenylate cyclase activating polypeptide-38 | | 著者 | Tateishi, Y, Jee, J.G, Inooka, H, Tochio, H, Hiroaki, H, Shirakawa, M. | | 登録日 | 2005-09-14 | | 公開日 | 2006-09-26 | | 最終更新日 | 2022-03-09 | | 実験手法 | SOLUTION NMR | | 主引用文献 | The solution structure of micelle-bound peptide
To be Published
|
|
6LFW
 
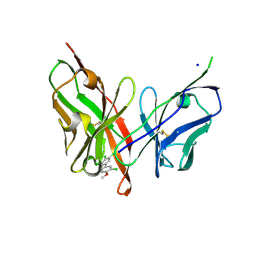 | |
6LCS
 
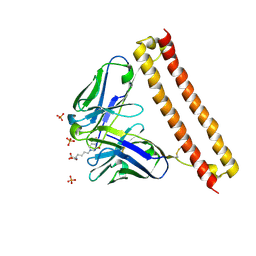 | | Crystal structure of 73MuL9 Fv-clasp fragment in complex with GA-pyridine analogue | | 分子名称: | (2~{S})-6-[4-(hydroxymethyl)-3-oxidanyl-pyridin-1-ium-1-yl]-2-(phenylmethoxycarbonylamino)hexanoic acid, PHOSPHATE ION, VH-SARAH, ... | | 著者 | Nakamura, T, Takagi, J, Yamagata, Y, Morioka, H. | | 登録日 | 2019-11-19 | | 公開日 | 2020-11-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Molecular recognition of a single-chain Fv antibody specific for GA-pyridine, an advanced glycation end-product (AGE), elucidated using biophysical techniques and synthetic antigen analogues.
J.Biochem., 170, 2021
|
|
6LFV
 
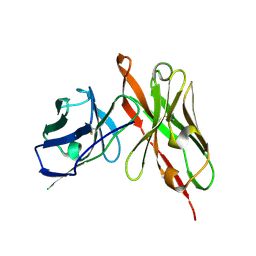 | |
2SSP
 
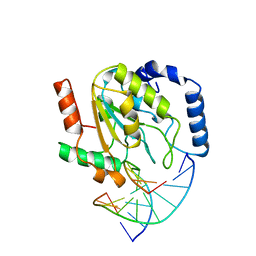 | | LEUCINE-272-ALANINE URACIL-DNA GLYCOSYLASE BOUND TO ABASIC SITE-CONTAINING DNA | | 分子名称: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*TP*(AAB)P*AP*TP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE) | | 著者 | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | 登録日 | 1999-04-28 | | 公開日 | 1999-05-06 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
2ZWS
 
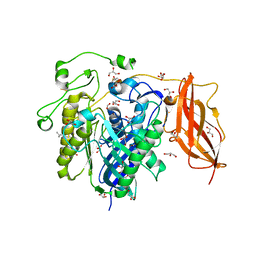 | | Crystal Structure Analysis of neutral ceramidase from Pseudomonas aeruginosa | | 分子名称: | FORMIC ACID, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Kakuta, Y, Okino, N, Inoue, T, Okano, H, Ito, M. | | 登録日 | 2008-12-17 | | 公開日 | 2009-03-03 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Mechanistic insights into the hydrolysis and synthesis of ceramide by neutral ceramidase.
J.Biol.Chem., 284, 2009
|
|
1ITY
 
 | | Solution structure of the DNA binding domain of human TRF1 | | 分子名称: | TRF1 | | 著者 | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-02-15 | | 公開日 | 2002-03-06 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a telomeric DNA complex of human TRF1
Structure, 9, 2001
|
|
1IV6
 
 | | Solution Structure of the DNA Complex of Human TRF1 | | 分子名称: | 5'-D(*CP*CP*CP*TP*AP*AP*CP*CP*CP*TP*AP*AP*C)-3', 5'-D(*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*G)-3', TELOMERIC REPEAT BINDING FACTOR 1 | | 著者 | Nishikawa, T, Okamura, H, Nagadoi, A, Konig, P, Rhodes, D, Nishimura, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | 登録日 | 2002-03-14 | | 公開日 | 2002-04-17 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a telomeric DNA complex of human TRF1.
Structure, 9, 2001
|
|
4BWS
 
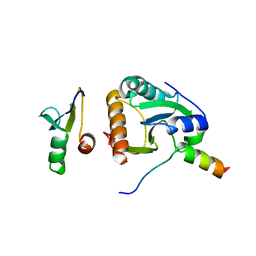 | | Crystal structure of the heterotrimer of PQBP1, U5-15kD and U5-52kD. | | 分子名称: | CD2 ANTIGEN CYTOPLASMIC TAIL-BINDING PROTEIN 2, POLYGLUTAMINE-BINDING PROTEIN 1, THIOREDOXIN-LIKE PROTEIN 4A | | 著者 | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Morimoto, T, Nabeshima, Y, Okazawa, H. | | 登録日 | 2013-07-04 | | 公開日 | 2014-04-30 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
1EMJ
 
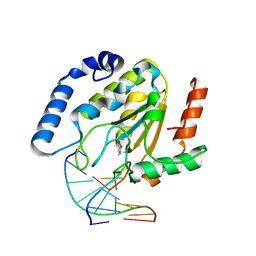 | | URACIL-DNA GLYCOSYLASE BOUND TO DNA CONTAINING A 4'-THIO-2'DEOXYURIDINE ANALOG PRODUCT | | 分子名称: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(ASU)P*AP*TP*CP*TP*T)-3'), URACIL, ... | | 著者 | Parikh, S.S, Walcher, G, Jones, G.D, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | 登録日 | 2000-03-16 | | 公開日 | 2000-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1UGH
 
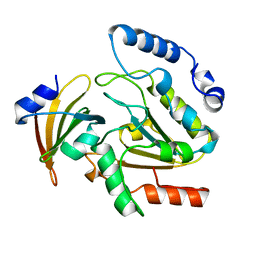 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE IN COMPLEX WITH A PROTEIN INHIBITOR: PROTEIN MIMICRY OF DNA | | 分子名称: | PROTEIN (URACIL-DNA GLYCOSYLASE INHIBITOR), PROTEIN (URACIL-DNA GLYCOSYLASE) | | 著者 | Mol, C.D, Arvai, A.S, Sanderson, R.J, Slupphaug, G, Kavli, B, Krokan, H.E, Mosbaugh, D.W, Tainer, J.A. | | 登録日 | 1999-02-05 | | 公開日 | 1999-02-16 | | 最終更新日 | 2023-08-23 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Crystal structure of human uracil-DNA glycosylase in complex with a protein inhibitor: protein mimicry of DNA.
Cell(Cambridge,Mass.), 82, 1995
|
|
1EMH
 
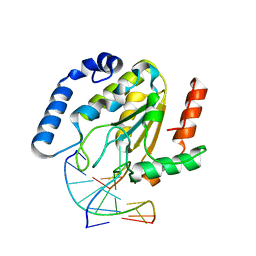 | | CRYSTAL STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO UNCLEAVED SUBSTRATE-CONTAINING DNA | | 分子名称: | DNA (5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*A)-3'), DNA (5'-D(*TP*GP*TP*(P2U)P*AP*TP*CP*TP*T)-3'), URACIL-DNA GLYCOSYLASE | | 著者 | Parikh, S.S, Slupphaug, G, Krokan, H.E, Blackburn, G.M, Tainer, J.A. | | 登録日 | 2000-03-16 | | 公開日 | 2000-05-16 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2RS2
 
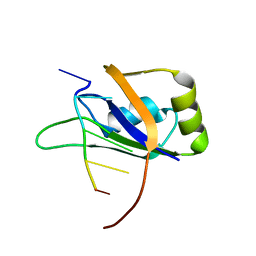 | | 1H, 13C, and 15N Chemical Shift Assignments for Musashi1 RBD1:r(GUAGU) complex | | 分子名称: | RNA (5'-R(*GP*UP*AP*GP*U)-3'), RNA-binding protein Musashi homolog 1 | | 著者 | Ohyama, T, Nagata, T, Tsuda, K, Imai, T, Okano, H, Yamazaki, T, Katahira, M. | | 登録日 | 2011-06-27 | | 公開日 | 2011-12-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure of Musashi1 in a complex with target RNA: the role of aromatic stacking interactions
Nucleic Acids Res., 2011
|
|
4SKN
 
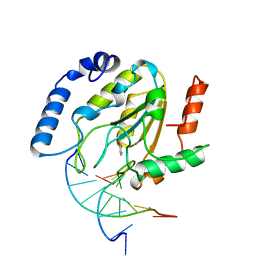 | | A NUCLEOTIDE-FLIPPING MECHANISM FROM THE STRUCTURE OF HUMAN URACIL-DNA GLYCOSYLASE BOUND TO DNA | | 分子名称: | DNA (5'-D(*AP*AP*AP*GP*CP*CP*GP*CP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*GP*(D1P)P*GP*GP*CP*TP*T)-3'), PROTEIN (URACIL-DNA GLYCOSYLASE), ... | | 著者 | Slupphaug, G, Mol, C.D, Kavli, B, Arvai, A.S, Krokan, H.E, Tainer, J.A. | | 登録日 | 1999-02-20 | | 公開日 | 1999-02-26 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | A nucleotide-flipping mechanism from the structure of human uracil-DNA glycosylase bound to DNA.
Nature, 384, 1996
|
|
2LQI
 
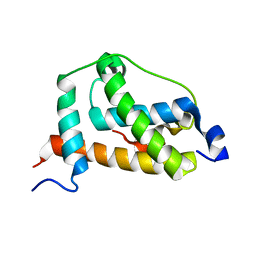 | | NMR structure of FOXO3a transactivation domains (CR2C-CR3) in complex with CBP KIX domain (2l3b conformation) | | 分子名称: | CREB-binding protein, Forkhead box O3 | | 著者 | Wang, F, Marshall, C.B, Yamamoto, K, Li, G.B, Gasmi-Seabrook, G.M.C, Okada, H, Mak, T.W, Ikura, M. | | 登録日 | 2012-03-06 | | 公開日 | 2012-05-16 | | 最終更新日 | 2024-05-15 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structures of KIX domain of CBP in complex with two FOXO3a transactivation domains reveal promiscuity and plasticity in coactivator recruitment.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1SSP
 
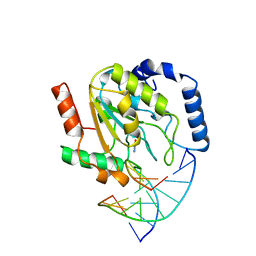 | | WILD-TYPE URACIL-DNA GLYCOSYLASE BOUND TO URACIL-CONTAINING DNA | | 分子名称: | 5'-D(*AP*AP*AP*GP*AP*TP*AP*AP*CP*AP*G)-3', 5'-D(*CP*TP*GP*TP*(D1P)P*AP*TP*CP*TP*T)-3', URACIL, ... | | 著者 | Parikh, S.S, Mol, C.D, Slupphaug, G, Bharati, S, Krokan, H.E, Tainer, J.A. | | 登録日 | 1999-04-28 | | 公開日 | 1999-05-06 | | 最終更新日 | 2023-08-02 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Base excision repair initiation revealed by crystal structures and binding kinetics of human uracil-DNA glycosylase with DNA.
EMBO J., 17, 1998
|
|
4CDO
 
 | | Crystal structure of PQBP1 bound to spliceosomal U5-15kD | | 分子名称: | THIOREDOXIN-LIKE PROTEIN 4A, POLYGLUTAMINE-BINDING PROTEIN | | 著者 | Mizuguchi, M, Obita, T, Serita, T, Kojima, R, Nabeshima, Y, Okazawa, H. | | 登録日 | 2013-11-05 | | 公開日 | 2014-04-30 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mutations in the Pqbp1 Gene Prevent its Interaction with the Spliceosomal Protein U5-15Kd.
Nat.Commun., 5, 2014
|
|
2Z33
 
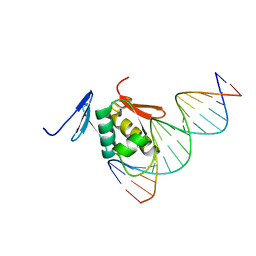 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | 分子名称: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | 著者 | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | 登録日 | 2007-05-31 | | 公開日 | 2008-04-22 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
