2YIH
 
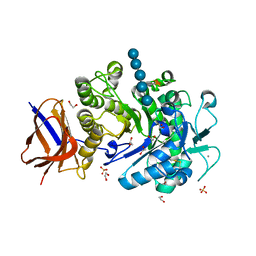 | | Structure of a Paenibacillus polymyxa Xyloglucanase from GH family 44 with Xyloglucan | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, CEL44C, ... | | Authors: | Ariza, A, Eklof, J.M, Spadiut, O, Offen, W.A, Roberts, S.M, Besenmatter, W, Friis, E.P, Skjot, M, Wilson, K.S, Brumer, H, Davies, G. | | Deposit date: | 2011-05-13 | | Release date: | 2011-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Activity of Paenibacillus Polymyxa Xyloglucanase from Glycoside Hydrolase Family 44.
J.Biol.Chem., 286, 2011
|
|
6ZBM
 
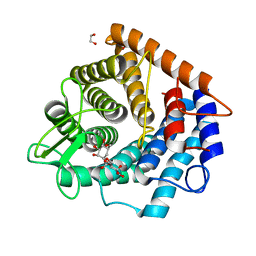 | | Structure of the D125N mutant of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol carbasugar-stabilised trisaccharide inhibitor | | Descriptor: | (1~{R},2~{R},3~{R},4~{S},5~{R})-4-[[(1~{S},2~{S},3~{S},4~{R},5~{R})-5-(hydroxymethyl)-2,3,4-tris(oxidanyl)cyclohexyl]oxymethyl]cyclohexane-1,2,3,5-tetrol, 1,2-ETHANEDIOL, Alpha-1,6-mannanase, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
6ZBX
 
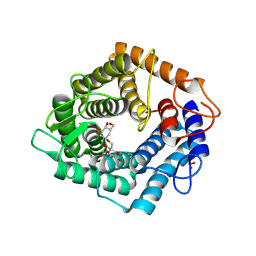 | | Structure of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6- alpha-manno-cyclophellitol carbasugar-stabilised trisaccharide inhibitor | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, ... | | Authors: | Schroeder, S, Offen, W.A, Jin, Y, De Boer, C, Enoterpi, J, Marino, L, van der Marel, G.A, Codee, J.D.C, Overkleeft, H.S, Davies, G.J. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
6ZBW
 
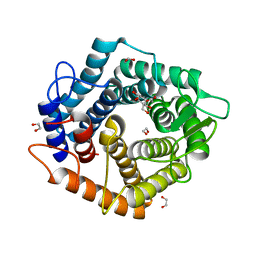 | | Structure of the D125N mutant of the catalytic domain of the Bacillus circulans alpha-1,6 Mannanase in complex with an alpha-1,6-alpha-manno-cyclophellitol trisaccharide inhibitor | | Descriptor: | (1~{S},4~{S},5~{R})-6-(hydroxymethyl)cyclohexane-1,2,3,4,5-pentol, 1,2-ETHANEDIOL, Alpha-1,6-mannanase, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2020-06-09 | | Release date: | 2021-04-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Development of Non-Hydrolysable Oligosaccharide Activity-Based Inactivators for Endoglycanases: A Case Study on alpha-1,6 Mannanases.
Chemistry, 27, 2021
|
|
5OYD
 
 | | GH5 endo-xyloglucanase from Cellvibrio japonicus | | Descriptor: | CHLORIDE ION, Cellulase, putative, ... | | Authors: | Attia, M, Nelson, C.E, Offen, W.A, Jain, N, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2017-09-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | In vitro and in vivo characterization of threeCellvibrio japonicusglycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions.
Biotechnol Biofuels, 11, 2018
|
|
5OYE
 
 | | GH5 endo-xyloglucanase from Cellvibrio japonicus | | Descriptor: | CHLORIDE ION, Cellulase, putative, ... | | Authors: | Attia, M, Nelson, C.E, Offen, W.A, Jain, N, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2017-09-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | In vitro and in vivo characterization of threeCellvibrio japonicusglycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions.
Biotechnol Biofuels, 11, 2018
|
|
5OST
 
 | | Beta-glucosidase from Thermoanaerobacterium xylolyticum GH116 in complex with Gluco-1H-imidazole | | Descriptor: | (4~{S},5~{S},6~{R},7~{R})-7-(hydroxymethyl)-4,5,6,7-tetrahydro-1~{H}-benzimidazole-4,5,6-triol, 1,2-ETHANEDIOL, CALCIUM ION, ... | | Authors: | Davies, G.J, Offen, W.A. | | Deposit date: | 2017-08-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gluco-1 H-imidazole: A New Class of Azole-Type beta-Glucosidase Inhibitor.
J. Am. Chem. Soc., 140, 2018
|
|
5OYC
 
 | | GH5 endo-xyloglucanase from Cellvibrio japonicus | | Descriptor: | CHLORIDE ION, Cellulase, putative, ... | | Authors: | Attia, M, Nelson, C.E, Offen, W.A, Jain, N, Gardner, J.G, Davies, G.J, Brumer, H. | | Deposit date: | 2017-09-08 | | Release date: | 2018-02-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | In vitro and in vivo characterization of threeCellvibrio japonicusglycoside hydrolase family 5 members reveals potent xyloglucan backbone-cleaving functions.
Biotechnol Biofuels, 11, 2018
|
|
6TEP
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6TEQ
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with 2-deoxy-2-fluoro-galactose | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-deoxy-2-fluoro-alpha-D-galactopyranose, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6TER
 
 | | Crystal structure of a galactokinase from Bifidobacterium infantis in complex with Galactose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Keenan, T, Parmeggiani, F, Fontenelle, C.Q, Malassis, J, Vendeville, J, Offen, W.A, Both, P, Huang, K, Marchesi, A, Heyam, A, Young, C, Charnock, S, Davies, G.J, Linclau, B, Flitsch, S.L, Fascione, M.A. | | Deposit date: | 2019-11-12 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Profiling Substrate Promiscuity of Wild-Type Sugar Kinases for Multi-fluorinated Monosaccharides.
Cell Chem Biol, 27, 2020
|
|
6I5Z
 
 | | Papaver somniferum O-methyltransferase | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, S-ADENOSYLMETHIONINE | | Authors: | Davies, G.J, Cabry, M.P, Offen, W.A. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
4BMX
 
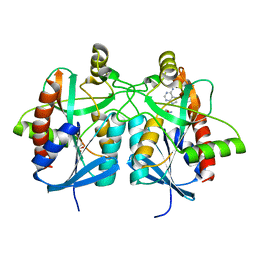 | | Native structure of futalosine hydrolase of Helicobacter pylori strain 26695 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENINE, MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4C7D
 
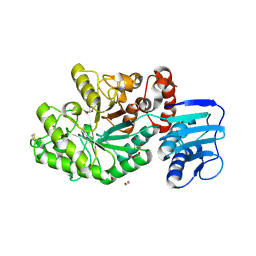 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
4C7G
 
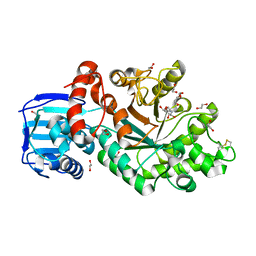 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
4BMZ
 
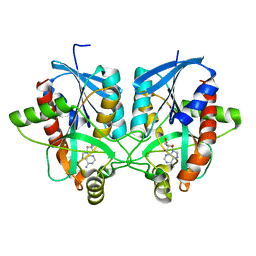 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6I6K
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
6I5Q
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | O-methyltransferase 1 | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-14 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
4C7F
 
 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, 6-ACETAMIDO-6-DEOXY-CASTANOSPERMINE, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyen Thi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
6I6L
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, Tetrahydrocolumbamine | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
4BMY
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | MTA/SAH NUCLEOSIDASE, SULFATE ION | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4BN0
 
 | | Structure of futalosine hydrolase mutant of Helicobacter pylori strain 26695 | | Descriptor: | MTA/SAH NUCLEOSIDASE | | Authors: | Kim, R.Q, Offen, W.A, Stubbs, K.A, Davies, G.J. | | Deposit date: | 2013-05-12 | | Release date: | 2013-09-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Structural Enzymology of Helicobacter Pylori Methylthioadenosine Nucleosidase in the Futalosine Pathway
Acta Crystallogr.,Sect.D, 70, 2014
|
|
6I6N
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
6I6M
 
 | | Papaver somniferum O-methyltransferase 1 | | Descriptor: | (13aS)-3,10-dimethoxy-5,8,13,13a-tetrahydro-6H-isoquino[3,2-a]isoquinoline-2,9-diol, O-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Cabry, M.P, Offen, W.A, Winzer, T, Li, Y, Graham, I.A, Davies, G.J, Saleh, P. | | Deposit date: | 2018-11-15 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structure of Papaver somniferum O-Methyltransferase 1 Reveals Initiation of Noscapine Biosynthesis with Implications for Plant Natural Product Methylation
Acs Catalysis, 2019
|
|
5M7R
 
 | | Structure of human O-GlcNAc hydrolase | | Descriptor: | Protein O-GlcNAcase | | Authors: | Roth, C, Chan, S, Offen, W.A, Hemsworth, G.R, Willems, L.I, King, D, Varghese, V, Britton, R, Vocadlo, D.J, Davies, G.J. | | Deposit date: | 2016-10-28 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional insight into human O-GlcNAcase.
Nat. Chem. Biol., 13, 2017
|
|
