7ABA
 
 | |
3ERK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE ERK2/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, EXTRACELLULAR REGULATED KINASE 2 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-09 | | Release date: | 1999-07-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
2X5N
 
 | | Crystal Structure of the SpRpn10 VWA domain | | Descriptor: | 26S PROTEASOME REGULATORY SUBUNIT RPN10, SULFATE ION | | Authors: | Riedinger, C, Boehringer, J, Trempe, J.-F, Lowe, E.D, Brown, N.R, Gehring, K, Noble, M.E.M, Gordon, C, Endicott, J.A. | | Deposit date: | 2010-02-10 | | Release date: | 2010-08-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The Structure of Rpn10 and its Interactions with Polyubiquitin Chains and the Proteasome Subunit Rpn12.
J.Biol.Chem., 285, 2010
|
|
6S10
 
 | |
6SAZ
 
 | | Cleaved human fetuin-b in complex with crayfish astacin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Astacin, ... | | Authors: | Gomis-Ruth, F.X, Guevara, T, Cuppari, A, Korschgen, H, Schmitz, C, Kuske, M, Yiallouros, I, Floehr, J, Jahnen-Dechent, W, Stocker, W. | | Deposit date: | 2019-07-18 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The C-terminal region of human plasma fetuin-B is dispensable for the raised-elephant-trunk mechanism of inhibition of astacin metallopeptidases.
Sci Rep, 9, 2019
|
|
3BL8
 
 | | Crystal structure of the extracellular domain of neuroligin 2A from mouse | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Neuroligin-2, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Jin, X, Koehnke, J, Shapiro, L. | | Deposit date: | 2007-12-10 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Crystal structure of the extracellular cholinesterase-like domain from neuroligin-2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3ELM
 
 | | Crystal Structure of MMP-13 Complexed with Inhibitor 24f | | Descriptor: | (2R)-({[5-(4-ethoxyphenyl)thiophen-2-yl]sulfonyl}amino){1-[(1-methylethoxy)carbonyl]piperidin-4-yl}ethanoic acid, CALCIUM ION, Collagenase 3, ... | | Authors: | Kulathila, R, Monovich, L, Koehn, J. | | Deposit date: | 2008-09-22 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of potent, selective, and orally active carboxylic acid based inhibitors of matrix metalloproteinase-13
J.Med.Chem., 52, 2009
|
|
6TET
 
 | | The structure of CYP121 in complex with inhibitor L21 | | Descriptor: | 1,2-ETHANEDIOL, 1-[(~{E})-3-[4-(4-fluorophenyl)phenyl]prop-2-enyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.49986887 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6TEV
 
 | | The structure of CYP121 in complex with inhibitor L44 | | Descriptor: | 1,2-ETHANEDIOL, 1-[[4-[4-(trifluoromethyl)phenyl]phenyl]methyl]imidazole, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-12 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.70001268 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6TE7
 
 | | The structure of CYP121 in complex with inhibitor S2 | | Descriptor: | 1,2-ETHANEDIOL, 2-chloranyl-4-[4-[(1~{R})-1-imidazol-1-ylprop-2-enyl]phenyl]phenol, Mycocyclosin synthase, ... | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2019-11-11 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.50001824 Å) | | Cite: | Structure-Activity Relationship and Mode-Of-Action Studies Highlight 1-(4-Biphenylylmethyl)-1H-imidazole-Derived Small Molecules as Potent CYP121 Inhibitors.
Chemmedchem, 16, 2021
|
|
6I86
 
 | | Crocagin biosynthetic gene J | | Descriptor: | GLYCEROL, Uncharacterized protein | | Authors: | Adam, S, Koehnke, J. | | Deposit date: | 2018-11-19 | | Release date: | 2019-03-13 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of CgnJ, a domain of unknown function protein from the crocagin gene cluster.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
3E3U
 
 | | Crystal structure of Mycobacterium tuberculosis peptide deformylase in complex with inhibitor | | Descriptor: | N-[(2R)-2-{[(2S)-2-(1,3-benzoxazol-2-yl)pyrrolidin-1-yl]carbonyl}hexyl]-N-hydroxyformamide, NICKEL (II) ION, Peptide deformylase | | Authors: | Meng, W, Xu, M, Pan, S, Koehn, J. | | Deposit date: | 2008-08-08 | | Release date: | 2009-01-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1B75
 
 | |
1BMK
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB218655 | | Descriptor: | 4-(FLUOROPHENYL)-1-CYCLOPROPYLMETHYL-5-(2-AMINO-4-PYRIMIDINYL)IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1A9U
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB203580 | | Descriptor: | 4-[5-(4-FLUORO-PHENYL)-2-(4-METHANESULFINYL-PHENYL)-3H-IMIDAZOL-4-YL]-PYRIDINE, MAP KINASE P38 | | Authors: | Wang, Z, Canagarajah, B, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-04-10 | | Release date: | 1999-04-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL6
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB216995 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-CYCLOROPROPYLMETHYL-5-(4-PYRIDYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-11 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
1BL7
 
 | | THE COMPLEX STRUCTURE OF THE MAP KINASE P38/SB220025 | | Descriptor: | 4-(4-FLUOROPHENYL)-1-(4-PIPERIDINYL)-5-(2-AMINO-4-PYRIMIDINYL)-IMIDAZOLE, PROTEIN (MAP KINASE P38) | | Authors: | Wang, Z, Canagarajah, B.J, Boehm, J.C, Kassis, S, Cobb, M.H, Young, P.R, Abdel-Meguid, S, Adams, J.L, Goldsmith, E.J. | | Deposit date: | 1998-07-23 | | Release date: | 1999-07-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of inhibitor selectivity in MAP kinases.
Structure, 6, 1998
|
|
5APG
 
 | | Structure of the SAM-dependent rRNA:acp-transferase Tsr3 from Vulcanisaeta distributa | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, TSR3, [(3S)-3-amino-4-hydroxy-4-oxo-butyl]-[[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-dihydroxy-oxolan-2-yl]methyl]-methyl-selanium | | Authors: | Wurm, J.P, Immer, C, Pogoryelov, D, Meyer, B, Koetter, P, Entian, K.-D, Woehnert, J. | | Deposit date: | 2015-09-15 | | Release date: | 2016-04-27 | | Last modified: | 2016-06-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ribosome Biogenesis Factor Tsr3 is the Aminocarboxypropyl Transferase Responsible for 18S Rrna Hypermodification in Yeast and Humans
Nucleic Acids Res., 44, 2016
|
|
5M9N
 
 | | Crystal structure of human TDRD1 extended Tudor domain in complex with a symmetrically dimethylated E2F peptide | | Descriptor: | 1,2-ETHANEDIOL, E2F peptide, N3, ... | | Authors: | Tallant, C, Savitsky, P, Moehlenbrink, J, Chan, C, Nunez-Alonso, G, Newman, J.A, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, La Thangue, N.B, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-01 | | Release date: | 2016-11-16 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human TDRD1 extended Tudor domain in complex with a symmetrically
dimethylated E2F peptide
To Be Published
|
|
5M9O
 
 | | Crystal structure of human SND1 extended Tudor domain in complex with a symmetrically dimethylated E2F peptide | | Descriptor: | E2F peptide, Staphylococcal nuclease domain-containing protein 1 | | Authors: | Tallant, C, Savitsky, P, Moehlenbrink, J, Chan, C, Nunez-Alonso, G, Siejka, P, Sorrell, F.J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Fedorov, O, La Thangue, N.B, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2016-11-01 | | Release date: | 2016-12-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of human SND1 extended Tudor domain in complex with a symmetrically
dimethylated E2F peptide
To Be Published
|
|
4BG2
 
 | | X-ray Crystal Structure of PatF from Prochloron didemni | | Descriptor: | PATF | | Authors: | Bent, A.F, Koehnke, J, Houssen, W.E, Smith, M.C.M, Jaspars, M, Naismith, J.H. | | Deposit date: | 2013-03-22 | | Release date: | 2013-04-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of Patf from Prochloron Didemni.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
2JQ7
 
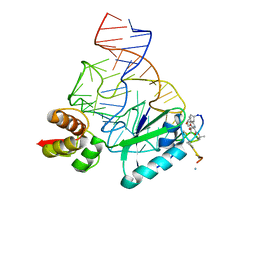 | | Model for thiostrepton binding to the ribosomal L11-RNA | | Descriptor: | 50S RIBOSOMAL PROTEIN L11, RIBOSOMAL RNA, THIOSTREPTON | | Authors: | Jonker, H.R.A, Ilin, S, Grimm, S.K, Woehnert, J, Schwalbe, H. | | Deposit date: | 2007-05-30 | | Release date: | 2007-07-03 | | Last modified: | 2021-08-18 | | Method: | SOLUTION NMR | | Cite: | L11 Domain Rearrangement Upon Binding to RNA and Thiostrepton Studied by NMR Spectroscopy
Nucleic Acids Res., 35, 2007
|
|
2LVL
 
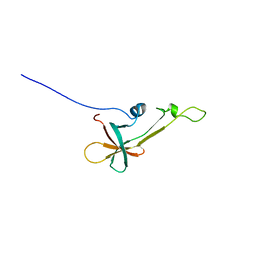 | | NMR Structure the lantibiotic immunity protein SpaI | | Descriptor: | SpaI | | Authors: | Christ, N, Bochmann, S, Gottstein, D, Duchardt-Ferner, E, Hellmich, U.A, Duesterhus, S, Koetter, P, Guentert, P, Entian, K, Woehnert, J. | | Deposit date: | 2012-07-06 | | Release date: | 2012-08-29 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The First Structure of a Lantibiotic Immunity Protein, SpaI from Bacillus subtilis, Reveals a Novel Fold.
J.Biol.Chem., 287, 2012
|
|
2MZJ
 
 | |
2KMJ
 
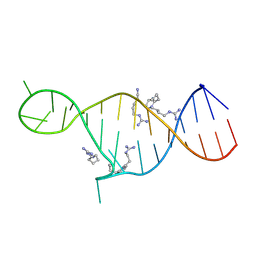 | | High resolution NMR solution structure of a complex of HIV-2 TAR RNA and a synthetic tripeptide in a 1:2 stoichiometry | | Descriptor: | Pyrimidinylpeptide, RNA (28-MER) | | Authors: | Ferner, J, Suhartono, M, Breitung, S, Jonker, H.R.A, Hennig, M, Woehnert, J, Goebel, M, Schwalbe, H. | | Deposit date: | 2009-07-30 | | Release date: | 2009-08-18 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Structures of HIV TAR RNA-ligand complexes reveal higher binding stoichiometries.
Chembiochem, 10, 2009
|
|
