1UBT
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
9AXU
 
 | | Non-translating S. pombe ribosome large subunit | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Gluc, M, Gemin, O, Purdy, M, Mattei, S, Jomaa, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (1.94 Å) | | Cite: | Ribosomes hibernate on mitochondria during cellular stress
Nat Commun, 15, 2024
|
|
9AXV
 
 | | Translating S. pombe ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Gluc, M, Gemin, O, Purdy, M, Mattei, S, Jomaa, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Ribosomes hibernate on mitochondria during cellular stress
Nat Commun, 15, 2024
|
|
9AXT
 
 | | Non-translating S. pombe ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Gluc, M, Gemin, O, Purdy, M, Mattei, S, Jomaa, A. | | Deposit date: | 2024-03-06 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Ribosomes hibernate on mitochondria during cellular stress
Nat Commun, 15, 2024
|
|
9DPF
 
 | | Crystal structure of TMPRSS11D S368A complexed with its own zymogen activation motif | | Descriptor: | 1,2-ETHANEDIOL, Transmembrane protease serine 11D, Transmembrane protease serine 11D non-catalytic chain | | Authors: | Fraser, B.J, Dong, A, Ilyassov, O, Seitova, A, Li, Y, Edwards, A, Benard, F, Arrowsmith, C, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-09-21 | | Release date: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of TMPRSS11D S368A complexed with its own zymogen activation motif
To Be Published
|
|
1UGV
 
 | | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621) | | Descriptor: | Olygophrenin-1 like protein | | Authors: | Inoue, K, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-20 | | Release date: | 2003-12-20 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SH3 domain of human olygophrein-1 like protein (KIAA0621)
To be Published
|
|
1TQR
 
 | | NMR Structure of DNA 17-mer GGAAAATCTCTAGCAGT corresponding to the extremity of the U5 LTR of the HIV-1 genome | | Descriptor: | DNA HIV-1 U5 LTR extremity | | Authors: | Renisio, J.G, Cosquer, S, Cherrak, I, El Antri, S, Mauffret, O, Fermandjian, S. | | Deposit date: | 2004-06-18 | | Release date: | 2005-04-19 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Pre-organized structure of viral DNA at the binding-processing site of HIV-1 integrase
NUCLEIC ACIDS RES., 33, 2005
|
|
9JR3
 
 | | Cryo-EM structure of PTH-PTH1R-Gq (tilted state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sano, F.K, Hirano, H, Itoh, Y, Nureki, O. | | Deposit date: | 2024-09-29 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Insights into G-protein coupling preference from cryo-EM structures of Gq-bound PTH1R
To Be Published
|
|
1OCA
 
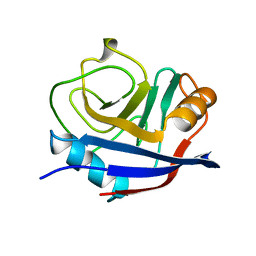 | | HUMAN CYCLOPHILIN A, UNLIGATED, NMR, 20 STRUCTURES | | Descriptor: | CYCLOPHILIN A | | Authors: | Ottiger, M, Zerbe, O, Guntert, P, Wuthrich, K. | | Deposit date: | 1997-07-07 | | Release date: | 1997-11-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The NMR solution conformation of unligated human cyclophilin A.
J.Mol.Biol., 272, 1997
|
|
1UBO
 
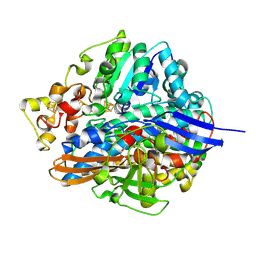 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1OQF
 
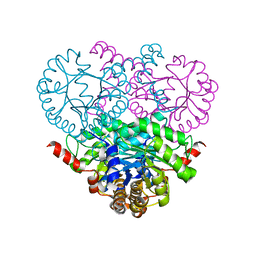 | | Crystal structure of the 2-methylisocitrate lyase | | Descriptor: | 2-methylisocitrate lyase | | Authors: | Liu, S, Lu, Z, Dunaway-Mariano, D, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-03-08 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structures of 2-methylisocitrate lyase in complex with product and with isocitrate inhibitor provide insight into lyase substrate specificity, catalysis and evolution.
Biochemistry, 44, 2005
|
|
1UDZ
 
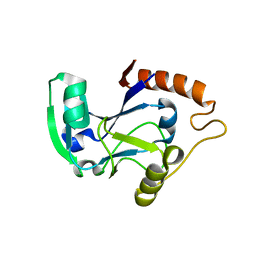 | | Isoleucyl-tRNA synthetase editing domain | | Descriptor: | Isoleucyl-tRNA synthetase | | Authors: | Fukunaga, R, Fukai, S, Ishitani, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-08 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine.
J.Biol.Chem., 279, 2004
|
|
1UGK
 
 | | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342) | | Descriptor: | Synaptotagmin IV | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the first C2 domain of synaptotagmin IV from human fetal brain (KIAA1342)
To be Published
|
|
1UDO
 
 | | Crystal structure of the tRNA processing enzyme RNase PH R86A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1OTV
 
 | | PqqC, Pyrroloquinolinquinone Synthase C | | Descriptor: | Coenzyme PQQ synthesis protein C | | Authors: | Magnusson, O.T, Toyama, H, Saeki, M, Rojas, A, Reed, J.C, Adachi, O, Klinman, J.P, SChwarzenbacher, R. | | Deposit date: | 2003-03-23 | | Release date: | 2004-05-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Quinone Biogenesis: Structure and Mechanism of PqqC, the Final Catalyst in the Production of Pyrroloquinoline Quinone.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
1P93
 
 | | CRYSTAL STRUCTURE OF THE AGONIST FORM OF GLUCOCORTICOID RECEPTOR | | Descriptor: | DEXAMETHASONE, Glucocorticoid receptor, Nuclear receptor coactivator 2 | | Authors: | Kauppi, B, Jakob, C, Farnegardh, M, Yang, J, Ahola, H, Alarcon, M, Calles, K, Engstrom, O, Harlan, J, Muchmore, S, Ramqvist, A.-K, Thorell, S, Ohman, L, Greer, J, Gustafsson, J.-A, Carlstedt-Duke, J, Carlquist, M. | | Deposit date: | 2003-05-09 | | Release date: | 2003-07-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Three-dimensional Structures of Antagonistic and Agonistic Forms of the Glucocorticoid Receptor Ligand-binding Domain:
RU-486 INDUCES A TRANSCONFORMATION THAT LEADS TO ACTIVE ANTAGONISM.
J.Biol.Chem., 278, 2003
|
|
1TZ5
 
 | | [pNPY19-23]-hPP bound to DPC Micelles | | Descriptor: | Pancreatic prohormone,neuropeptide Y,Pancreatic prohormone | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
1UBL
 
 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
1UAG
 
 | | UDP-N-ACETYLMURAMOYL-L-ALANINE:D-GLUTAMATE LIGASE | | Descriptor: | SULFATE ION, UDP-N-ACETYLMURAMOYL-L-ALANINE/:D-GLUTAMATE LIGASE, URIDINE-5'-DIPHOSPHATE-N-ACETYLMURAMOYL-L-ALANINE | | Authors: | Bertrand, J, Fanchon, E, Dideberg, O. | | Deposit date: | 1997-03-13 | | Release date: | 1998-03-18 | | Last modified: | 2018-04-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of UDP-N-acetylmuramoyl-L-alanine:D-glutamate ligase from Escherichia coli.
EMBO J., 16, 1997
|
|
1UDQ
 
 | | Crystal structure of the tRNA processing enzyme RNase PH T125A mutant from Aquifex aeolicus | | Descriptor: | PHOSPHATE ION, Ribonuclease PH, SULFATE ION | | Authors: | Ishii, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-02 | | Release date: | 2003-09-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the tRNA Processing Enzyme RNase PH from Aquifex aeolicus
J.Biol.Chem., 278, 2003
|
|
1UE0
 
 | | Isoleucyl-tRNA synthetase editing domain complexed with L-Valine | | Descriptor: | Isoleucyl-tRNA synthetase, VALINE | | Authors: | Fukunaga, R, Fukai, S, Ishitani, R, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-08 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of the CP1 domain from Thermus thermophilus isoleucyl-tRNA synthetase and its complex with L-valine.
J.Biol.Chem., 279, 2004
|
|
1UFX
 
 | | Solution structure of the third PDZ domain of human KIAA1526 protein | | Descriptor: | KIAA1526 protein | | Authors: | Tochio, N, Kobayashi, N, Koshiba, S, Kigawa, T, Inoue, M, Shirouzu, M, Terada, T, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Matsuo, Y, Ohara, O, Nagase, T, Kikuno, R, Nakayama, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-10 | | Release date: | 2003-12-10 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the third PDZ domain of human KIAA1526 protein
To be Published
|
|
1UEU
 
 | |
1NNX
 
 | | Structure of the hypothetical protein ygiW from E. coli. | | Descriptor: | Protein ygiW, SULFATE ION | | Authors: | Lehmann, C, Galkin, A, Pullalarevu, S, Sarikaya, E, Krajewski, W, Lim, K, Howard, A, Herzberg, O, Structure 2 Function Project (S2F) | | Deposit date: | 2003-01-14 | | Release date: | 2004-03-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure of the hypothetical protein ygiW from E. coli.
To be Published
|
|
8CUD
 
 | |
