2YDX
 
 | | Crystal structure of human S-adenosylmethionine synthetase 2, beta subunit | | Descriptor: | 1,4,5,6-TETRAHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE PHOSPHATE, CALCIUM ION, METHIONINE ADENOSYLTRANSFERASE 2 SUBUNIT BETA, ... | | Authors: | Muniz, J.R.C, Shafqat, N, Pike, A.C.W, Yue, W.W, Vollmar, M, Papagriogriou, V, Roos, A, Gileadi, O, von Delft, F, Kavanagh, K.L, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Oppermann, U. | | Deposit date: | 2011-03-25 | | Release date: | 2011-04-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight Into S-Adenosylmethionine Biosynthesis from the Crystal Structures of the Human Methionine Adenosyltransferase Catalytic and Regulatory Subunits.
Biochem.J., 452, 2013
|
|
3P1D
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with N-Methyl-2-pyrrolidone (NMP) | | Descriptor: | 1-methylpyrrolidin-2-one, CREB-binding protein, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3P1C
 
 | | Crystal structure of the bromodomain of human CREBBP in complex with acetylated lysine | | Descriptor: | CREB-binding protein, N(6)-ACETYLLYSINE, POTASSIUM ION, ... | | Authors: | Filippakopoulos, P, Picaud, S, Feletar, I, Fedorov, O, Muniz, J, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-09-30 | | Release date: | 2010-11-24 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
2WO6
 
 | | Human Dual-Specificity Tyrosine-Phosphorylation-Regulated Kinase 1A in complex with a consensus substrate peptide | | Descriptor: | ARTIFICIAL CONSENSUS SEQUENCE, CHLORIDE ION, DUAL SPECIFICITY TYROSINE-PHOSPHORYLATION- REGULATED KINASE 1A, ... | | Authors: | Roos, A.K, Soundararajan, M, Elkins, J.M, Fedorov, O, Eswaran, J, Phillips, C, Pike, A.C.W, Ugochukwu, E, Muniz, J.R.C, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Wikstrom, M, Edwards, A, Bountra, C, Knapp, S. | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Down Syndrome Kinases, Dyrks, Reveal Mechanisms of Kinase Activation and Substrate Recognition.
Structure, 21, 2013
|
|
2XJJ
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1,3-DIHYDROISOINDOL-2-YL-(6-HYDROXY-3,3-DIMETHYL-1,2-DIHYDROINDOL-5-YL)METHANONE, GLYCEROL, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2X6S
 
 | | Human foamy virus integrase - catalytic core. Magnesium-bound structure. | | Descriptor: | INTEGRASE, MAGNESIUM ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-19 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
2X6N
 
 | | Human foamy virus integrase - catalytic core. Manganese-bound structure. | | Descriptor: | INTEGRASE, MANGANESE (II) ION | | Authors: | Rety, S, Delelis, O, Rezabkova, L, Dubanchet, B, Silhan, J, Lewit-Bentley, A. | | Deposit date: | 2010-02-18 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structural Studies of the Catalytic Core of the Primate Foamy Virus (Pfv-1) Integrase
Acta Crystallogr.,Sect.F, 66, 2010
|
|
3QPI
 
 | | Crystal Structure of Dimeric Chlorite Dismutases from Nitrobacter winogradskyi | | Descriptor: | Chlorite Dismutase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mlynek, G, Sjoeblom, B, Kostan, J, Fuereder, S, Maixner, F, Furtmueller, P.G, Obinger, O, Wagner, M, Daims, H, Djinovic-Carugo, K. | | Deposit date: | 2011-02-13 | | Release date: | 2011-07-06 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Unexpected diversity of chlorite dismutases: a catalytically efficient dimeric enzyme from Nitrobacter winogradskyi.
J.Bacteriol., 193, 2011
|
|
2XJG
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | 1,3-DIHYDROISOINDOL-2-YL-(2-HYDROXY-4-METHOXY-5-PROPAN-2-YL-PHENYL)METHANONE, HEAT SHOCK PROTEIN HSP 90-ALPHA | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, B, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
3Q4S
 
 | | Crystal Structure of Human Glycogenin-1 (GYG1), apo form | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Glycogenin-1 | | Authors: | Chaikuad, A, Froese, D.S, Yue, W.W, Krysztofinska, E, von Delft, F, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Oppermann, O, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-24 | | Release date: | 2011-02-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Conformational plasticity of glycogenin and its maltosaccharide substrate during glycogen biogenesis.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3Q2E
 
 | | Crystal structure of the second bromodomain of human bromodomain and WD repeat-containing protein 1 isoform A (WDR9) | | Descriptor: | ACETATE ION, Bromodomain and WD repeat-containing protein 1 | | Authors: | Filippakopoulos, P, Felletar, I, Picaud, S, Keates, T, Krojer, T, Muniz, J, Gileadi, O, Von Delft, F, Arrowsmith, C.H, Edwards, A, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Histone recognition and large-scale structural analysis of the human bromodomain family.
Cell(Cambridge,Mass.), 149, 2012
|
|
3OP4
 
 | | Crystal structure of putative 3-ketoacyl-(acyl-carrier-protein) reductase from Vibrio cholerae O1 biovar eltor str. N16961 in complex with NADP+ | | Descriptor: | 3-oxoacyl-[acyl-carrier protein] reductase, ACETATE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hou, J, Chruszcz, M, Onopriyenko, O, Grimshaw, S, Porebski, P, Zheng, H, Savchenko, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-31 | | Release date: | 2010-09-22 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Dissecting the Structural Elements for the Activation of beta-Ketoacyl-(Acyl Carrier Protein) Reductase from Vibrio cholerae.
J.Bacteriol., 198, 2015
|
|
2XZR
 
 | | Escherichia coli Immunoglobulin-binding protein EibD 391-438 FUSED TO GCN4 ADAPTORS | | Descriptor: | CHLORIDE ION, IMMUNOGLOBULIN-BINDING PROTEIN EIBD | | Authors: | Hartmann, M.D, Hernandez Alvarez, B, Ridderbusch, O, Deiss, S, Lupas, A.N. | | Deposit date: | 2010-11-28 | | Release date: | 2011-07-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Structure of E. Coli Igg-Binding Protein D Suggests a General Model for Bending and Binding in Trimeric Autotransporter Adhesins.
Structure, 19, 2011
|
|
2XAB
 
 | | Structure of HSP90 with an inhibitor bound | | Descriptor: | 4-(1,3-DIHYDRO-2H-ISOINDOL-2-YLCARBONYL)-6-(1-METHYLETHYL)BENZENE-1,3-DIOL, HEAT SHOCK PROTEIN HSP 90-ALPHA, | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, m, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, O'Brian, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-03-30 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2XJX
 
 | | Structure of HSP90 with small molecule inhibitor bound | | Descriptor: | HEAT SHOCK PROTEIN HSP 90-ALPHA, Onalespib | | Authors: | Murray, C.W, Carr, M.G, Callaghan, O, Chessari, G, Congreve, M, Cowan, S, Coyle, J.E, Downham, R, Figueroa, E, Frederickson, M, Graham, B, McMenamin, R, OBrien, M.A, Patel, S, Phillips, T.R, Williams, G, Woodhead, A.J, Woolford, A.J.A. | | Deposit date: | 2010-07-06 | | Release date: | 2010-08-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Discovery of (2,4-Dihydroxy-5-Isopropylphenyl)-[5-(4-Methylpiperazin-1-Ylmethyl)-1,3-Dihydroisoindol-2-Yl]Methanone (at13387), a Novel Inhibitor of the Molecular Chaperone Hsp90 by Fragment Based Drug Design.
J.Med.Chem., 53, 2010
|
|
2ZQQ
 
 | | Crystal structure of human AUH (3-methylglutaconyl-coa hydratase) mixed with (AUUU)24A RNA | | Descriptor: | Methylglutaconyl-CoA hydratase | | Authors: | Kurimoto, K, Kuwasako, K, Muto, Y, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | AU-rich RNA-binding induces changes in the quaternary structure of AUH
Proteins, 75, 2009
|
|
3NX3
 
 | | Crystal structure of acetylornithine aminotransferase (argD) from Campylobacter jejuni | | Descriptor: | Acetylornithine aminotransferase, MAGNESIUM ION | | Authors: | Anderson, S.M, Wawrzak, Z, Onopriyenko, O, Skarina, T, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-12 | | Release date: | 2010-08-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: |
|
|
3B7O
 
 | | Crystal structure of the human tyrosine phosphatase SHP2 (PTPN11) with an accessible active site | | Descriptor: | D-MALATE, Tyrosine-protein phosphatase non-receptor type 11 | | Authors: | Ugochukwu, E, Barr, A, Patel, A, King, O, Niesen, F, Salah, E, Savitsky, P, Pilka, E.S, Elkins, J, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, von Delft, F, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-10-31 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Large-scale structural analysis of the classical human protein tyrosine phosphatome.
Cell(Cambridge,Mass.), 136, 2009
|
|
2ZQR
 
 | | Crystal structure of AUH without RNA | | Descriptor: | Methylglutaconyl-CoA hydratase | | Authors: | Kurimoto, K, Kuwasako, K, Muto, Y, Nureki, O, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2008-08-16 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | AU-rich RNA-binding induces changes in the quaternary structure of AUH
Proteins, 75, 2009
|
|
3BIC
 
 | | Crystal structure of human methylmalonyl-CoA mutase | | Descriptor: | CHLORIDE ION, Methylmalonyl-CoA mutase, mitochondrial precursor | | Authors: | Ugochukwu, E, Kochan, G, Pantic, N, Parizotto, E, Pilka, E.S, Pike, A.C.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-11-30 | | Release date: | 2007-12-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of the human GTPase MMAA and vitamin B12-dependent methylmalonyl-CoA mutase and insight into their complex formation.
J.Biol.Chem., 285, 2010
|
|
6QH1
 
 | | The structure of Schizosaccharomyces pombe PCNA in complex with an Spd1 derived peptide | | Descriptor: | Proliferating cell nuclear antigen, S-phase delaying protein 1 | | Authors: | Kragelund, B.B, Nielsen, O, Olsen, J.G, Kassem, N, Prestel, A. | | Deposit date: | 2019-01-14 | | Release date: | 2020-02-05 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | The structure of Schizosaccharomyces pombe PCNA in complex with an Spd1 derived peptide
To Be Published
|
|
6R63
 
 | | Crystal structure of indoleamine 2,3-dioxygenase 1 (IDO1) in complex with ferric heme and MMG-0358 | | Descriptor: | 4-chloranyl-2-(2~{H}-1,2,3-triazol-4-yl)phenol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Roehrig, U.F, Reynaud, A, Pojer, F, Michielin, O, Zoete, V. | | Deposit date: | 2019-03-26 | | Release date: | 2019-10-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.894 Å) | | Cite: | Inhibition Mechanisms of Indoleamine 2,3-Dioxygenase 1 (IDO1).
J.Med.Chem., 62, 2019
|
|
6R65
 
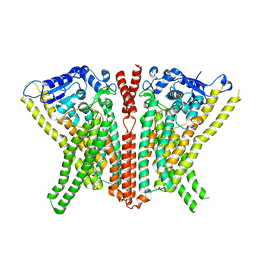 | | Crystal Structure of human TMEM16K / Anoctamin 10 (Form 2) | | Descriptor: | Anoctamin-10, CALCIUM ION | | Authors: | Bushell, S.R, Pike, A.C.W, Chu, A, Tessitore, A, Rotty, B, Mukhopadhyay, S, Kupinska, K, Shrestha, L, Borkowska, O, Chalk, R, Burgess-Brown, N.A, Love, J, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-03-26 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The structural basis of lipid scrambling and inactivation in the endoplasmic reticulum scramblase TMEM16K.
Nat Commun, 10, 2019
|
|
6S3Q
 
 | | Structure of human excitatory amino acid transporter 3 (EAAT3) in complex with TFB-TBOA | | Descriptor: | (2~{S},3~{S})-2-azanyl-3-[[3-[[4-(trifluoromethyl)phenyl]carbonylamino]phenyl]methoxy]butanedioic acid, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Baronina, A, Pike, A.C.W, Yu, X, Dong, Y.Y, Shintre, C.A, Tessitore, A, Chu, A, Rotty, B, Venkaya, S, Mukhopadhyay, S, Borkowska, O, Chalk, R, Shrestha, L, Burgess-Brown, N.A, Edwards, A.M, Arrowsmith, C.H, Bountra, C, Han, S, Carpenter, E.P, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-25 | | Release date: | 2020-07-08 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structure of human excitatory amino acid transporter 3 (EAAT3)
TO BE PUBLISHED
|
|
6QY6
 
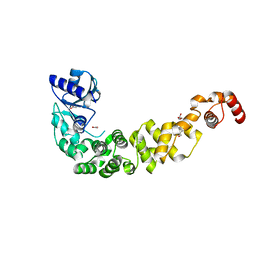 | | Crystal structure of the CCA-adding enzyme of a psychrophilic organism | | Descriptor: | ACETATE ION, CCA-adding enzyme, GLYCEROL, ... | | Authors: | de Wijn, R, Rollet, K, Bluhm, A, Hennig, O, Betat, H, Moerl, M, Lorber, B, Sauter, C. | | Deposit date: | 2019-03-08 | | Release date: | 2020-03-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CCA-addition in the cold: Structural characterization of the psychrophilic CCA-adding enzyme from the permafrost bacterium Planococcus halocryophilus
Comput Struct Biotechnol J, 2021
|
|
