1UUU
 
 | | STRUCTURE OF AN RNA HAIRPIN LOOP WITH A 5'-CGUUUCG-3' LOOP MOTIF BY HETERONUCLEAR NMR SPECTROSCOPY AND DISTANCE GEOMETRY, 15 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*GP*UP*AP*CP*GP*UP*UP*UP*CP*GP*UP*AP*CP*GP*CP*C)-3') | | Authors: | Sich, C, Ohlenschlager, O, Ramachandran, R, Gorlach, M, Brown, L.R. | | Deposit date: | 1997-08-12 | | Release date: | 1998-02-25 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of an RNA hairpin loop with a 5'-CGUUUCG-3' loop motif by heteronuclear NMR spectroscopy and distance geometry.
Biochemistry, 36, 1997
|
|
1V34
 
 | | Crystal structure of Pyrococcus horikoshii DNA primase-UTP complex | | Descriptor: | DNA primase small subunit, URIDINE 5'-TRIPHOSPHATE, ZINC ION | | Authors: | Ito, N, Nureki, O, Shirouzu, M, Yokoyama, S, Hanaoka, F, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-10-25 | | Release date: | 2004-03-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of the Pyrococcus horikoshii DNA primase-UTP complex: implications for the mechanism of primer synthesis.
Genes Cells, 8, 2003
|
|
1UPT
 
 | | Structure of a complex of the golgin-245 GRIP domain with Arl1 | | Descriptor: | ADP-RIBOSYLATION FACTOR-LIKE PROTEIN 1, GOLGI AUTOANTIGEN, GOLGIN SUBFAMILY A MEMBER 4, ... | | Authors: | Panic, B, Perisic, O, Veprintsev, D.B, Williams, R.L, Munro, S. | | Deposit date: | 2003-10-12 | | Release date: | 2003-10-30 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Arl1-Dependent Targeting of Homodimeric Grip Domains to the Golgi Apparatus
Mol.Cell, 12, 2003
|
|
1UZC
 
 | | THE STRUCTURE OF AN FF DOMAIN FROM HUMAN HYPA/FBP11 | | Descriptor: | HYPOTHETICAL PROTEIN FLJ21157 | | Authors: | Allen, M.D, Jemth, P, Friedler, A, Schon, O, Bycroft, M. | | Deposit date: | 2004-03-09 | | Release date: | 2004-04-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Structure of an Ff Domain from Human Hypa/Fbp11.
J.Mol.Biol., 323, 2002
|
|
1VAV
 
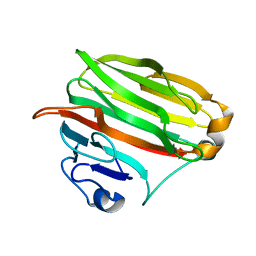 | | Crystal structure of alginate lyase PA1167 from Pseudomonas aeruginosa at 2.0 A resolution | | Descriptor: | Alginate lyase PA1167 | | Authors: | Yamasaki, M, Moriwaki, S, Miyake, O, Hashimoto, W, Murata, K, Mikami, B. | | Deposit date: | 2004-02-19 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and function of a hypothetical Pseudomonas aeruginosa protein PA1167 classified into family PL-7: a novel alginate lyase with a beta-sandwich fold.
J.Biol.Chem., 279, 2004
|
|
5JJV
 
 | |
4XJ3
 
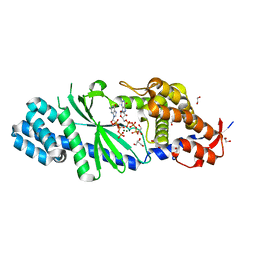 | | Crystal structure of Vibrio cholerae DncV GTP bound form | | Descriptor: | 1,2-ETHANEDIOL, Cyclic AMP-GMP synthase, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Kato, K, Ishii, R, Ishitani, R, Nureki, O. | | Deposit date: | 2015-01-08 | | Release date: | 2015-04-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Basis for the Catalytic Mechanism of DncV, Bacterial Homolog of Cyclic GMP-AMP Synthase
Structure, 23, 2015
|
|
5JOE
 
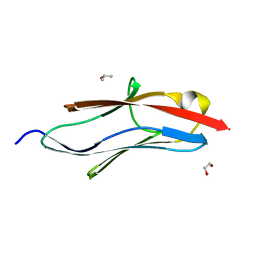 | | Crystal structure of I81 from titin | | Descriptor: | 1,2-ETHANEDIOL, ISOPROPYL ALCOHOL, Titin | | Authors: | Fleming, J, Zhou, T, Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CARP interacts with titin at a unique helical N2A sequence and at the domain Ig81 to form a structured complex.
Febs Lett., 590, 2016
|
|
5JOM
 
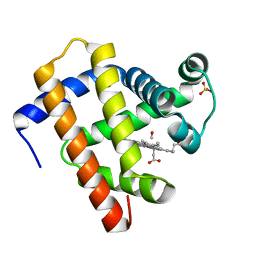 | | X-ray structure of CO-bound sperm whale myoglobin using a fixed target crystallography chip | | Descriptor: | CARBON MONOXIDE, Myoglobin, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Oghbaey, S, Sarracini, A, Ginn, H.M, Pare-Labrosse, O, Kuo, A, Marx, A, Epp, S.W, Sherrell, D.A, Eger, B.T, Zhong, Y, Loch, R, Mariani, V, Alonso-Mori, R, Nelson, S, Lemke, H.T, Owen, R.L, Pearson, A.R, Stuart, D.I, Ernst, O.P, Mueller-Werkmeister, H.M, Miller, R.J.D. | | Deposit date: | 2016-05-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fixed target combined with spectral mapping: approaching 100% hit rates for serial crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
6FWG
 
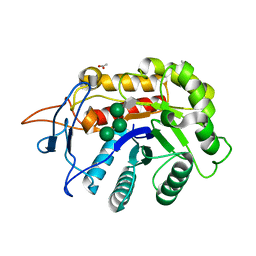 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with tetramannoside yeast mannan fragment | | Descriptor: | ACETATE ION, Glycosyl hydrolase family 71, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-methyl alpha-D-mannopyranoside | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6FWM
 
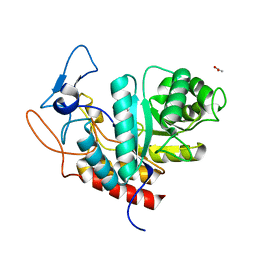 | | Structure of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-Glc-1,3-1,2-anhydro-mannose hydrolyzed by enzyme | | Descriptor: | (1~{S},2~{R},3~{S},4~{R},5~{R})-5-(hydroxymethyl)cyclohexane-1,2,3,4-tetrol, ACETATE ION, Glycosyl hydrolase family 71, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-06 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
6FWQ
 
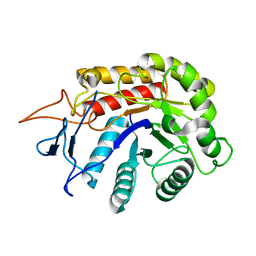 | | Structure of an E333Q variant of the GH99 endo-alpha-mannanase from Bacteroides xylanisolvens in complex with alpha-1,3-mannobiose and alpha-1,2-mannobiose | | Descriptor: | Glycosyl hydrolase family 71, SODIUM ION, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose, ... | | Authors: | Sobala, L.F, Speciale, G, Hakki, Z, Fernandes, P.Z, Raich, L, Rojas-Cervellera, V, Bennet, A, Thompson, A.J, Bernardo-Seisdedos, G, Millet, O, Zhu, S, Lu, D, Sollogoub, M, Rovira, C, Jimenez-Barbero, J, Davies, G.J, Williams, S.J. | | Deposit date: | 2018-03-07 | | Release date: | 2019-09-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Epoxide Intermediate in Glycosidase Catalysis.
Acs Cent.Sci., 6, 2020
|
|
4E5B
 
 | |
4EHI
 
 | | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Bifunctional purine biosynthesis protein PurH, SULFATE ION | | Authors: | Brunzelle, J.S, Wawrzak, Z, Onopriyenko, O, Kwok, J, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-06-06 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | An X-ray Crystal Structure of a putative Bifunctional Phosphoribosylaminoimidazolecarboxamide Formyltransferase/IMP Cyclohydrolase
TO BE PUBLISHED
|
|
5JOW
 
 | | Bacteroides ovatus Xyloglucan PUL GH43A | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Non-reducing end alpha-L-arabinofuranosidase BoGH43A | | Authors: | Thompson, A.J, Hemsworth, G.R, Stepper, J, Sobala, L.F, Coyle, T, Larsbrink, J, Spadiut, O, Stubbs, K.A, Brumer, H, Davies, G.J. | | Deposit date: | 2016-05-03 | | Release date: | 2016-08-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural dissection of a complex Bacteroides ovatus gene locus conferring xyloglucan metabolism in the human gut.
Open Biology, 6, 2016
|
|
2FK6
 
 | | Crystal Structure of RNAse Z/tRNA(Thr) complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, RIBONUCLEASE Z, ... | | Authors: | Li de la Sierra-Gallay, I, Mathy, N, Pellegrini, O, Condon, C. | | Deposit date: | 2006-01-04 | | Release date: | 2006-03-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the ubiquitous 3' processing enzyme RNase Z bound to transfer RNA.
Nat.Struct.Mol.Biol., 13, 2006
|
|
4EJ7
 
 | | Crystal structure of the aminoglycoside phosphotransferase APH(3')-Ia, ATP-bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Aminoglycoside 3'-phosphotransferase AphA1-IAB, CALCIUM ION, ... | | Authors: | Stogios, P.J, Minasov, G, Tan, K, Evdokimova, E, Egorova, O, Di Leo, R, Shakya, T, Wright, G.D, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-06 | | Release date: | 2012-04-18 | | Last modified: | 2013-09-04 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure-guided optimization of protein kinase inhibitors reverses aminoglycoside antibiotic resistance.
Biochem.J., 454, 2013
|
|
4XIY
 
 | | Crystal structure of ketol-acid reductoisomerase from Azotobacter | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Ketol-acid reductoisomerase, ... | | Authors: | Spatzal, T, Cahn, J.K.B, Wiig, J.A, Einsle, O, Hu, Y, Ribbe, M.W, Arnold, F.H. | | Deposit date: | 2015-01-07 | | Release date: | 2015-04-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cofactor specificity motifs and the induced fit mechanism in class I ketol-acid reductoisomerases.
Biochem.J., 468, 2015
|
|
5KC2
 
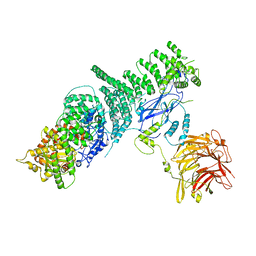 | | Negative stain structure of Vps15/Vps34 complex | | Descriptor: | Phosphatidylinositol 3-kinase VPS34, Serine/threonine-protein kinase VPS15 | | Authors: | Kirsten, M.L, Zhang, L, Ohashi, Y, Perisic, O, Williams, R.L, Sachse, C. | | Deposit date: | 2016-06-04 | | Release date: | 2016-10-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Characterization of Atg38 and NRBF2, a fifth subunit of the autophagic Vps34/PIK3C3 complex.
Autophagy, 12, 2016
|
|
4IAC
 
 | |
4IAI
 
 | |
4IB3
 
 | |
4I22
 
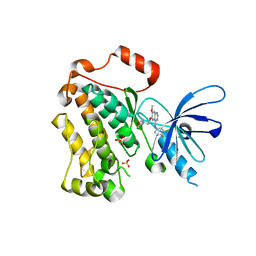 | | Structure of the monomeric (V948R)gefitinib/erlotinib resistant double mutant (L858R+T790M) EGFR kinase domain co-crystallized with gefitinib | | Descriptor: | Epidermal growth factor receptor, Gefitinib, SULFATE ION | | Authors: | Gajiwala, K.S, Feng, J, Ferre, R, Ryan, K, Brodsky, O, Stewart, A. | | Deposit date: | 2012-11-21 | | Release date: | 2013-01-16 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Insights into the Aberrant Activity of Mutant EGFR Kinase Domain and Drug Recognition.
Structure, 21, 2013
|
|
5JPQ
 
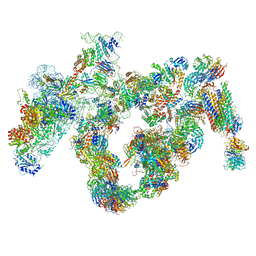 | | Cryo-EM structure of the 90S pre-ribosome | | Descriptor: | 18S ribosomal RNA, Bms1, Emg1, ... | | Authors: | Turk, M, Cheng, J, Berninghausen, O, Kornprobst, M, Flemming, D, Kos-Braun, I.C, Kos, M, Thoms, M, Hurt, E, Beckmann, R. | | Deposit date: | 2016-05-04 | | Release date: | 2016-07-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.3 Å) | | Cite: | Architecture of the 90S Pre-ribosome: A Structural View on the Birth of the Eukaryotic Ribosome.
Cell, 166, 2016
|
|
4XW6
 
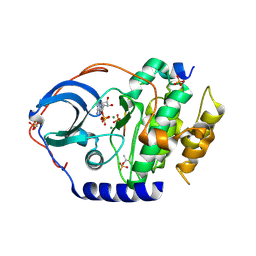 | | X-ray structure of PKAc with ADP, free phosphate ion, CP20, magnesium ions | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Gerlits, O, Tian, J, Das, A, Taylor, S, Langan, P, Heller, T.W, Kovalevsky, A. | | Deposit date: | 2015-01-28 | | Release date: | 2015-05-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphoryl Transfer Reaction Snapshots in Crystals: INSIGHTS INTO THE MECHANISM OF PROTEIN KINASE A CATALYTIC SUBUNIT.
J.Biol.Chem., 290, 2015
|
|
