3CF6
 
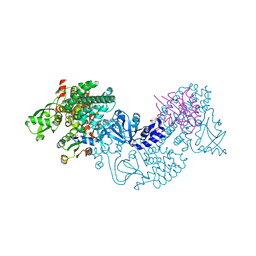 | | Structure of Epac2 in complex with cyclic-AMP and Rap | | Descriptor: | 6-(6-AMINO-PURIN-9-YL)-2-THIOXO-TETRAHYDRO-2-FURO[3,2-D][1,3,2]DIOXAPHOSPHININE-2,7-DIOL, Rap guanine nucleotide exchange factor (GEF) 4, Ras-related protein Rap-1b, ... | | Authors: | Rehmann, H, Arias-Palomo, E, Hadders, M.A, Schwede, F, Llorca, O, Bos, J.L. | | Deposit date: | 2008-03-02 | | Release date: | 2008-07-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of Epac2 in complex with a cyclic AMP analogue and RAP1B
Nature, 455, 2008
|
|
3CPJ
 
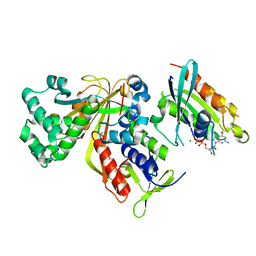 | | Crystal structure of Ypt31 in complex with yeast Rab-GDI | | Descriptor: | CHLORIDE ION, GTP-binding protein YPT31/YPT8, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Kravchenko, S, Ignatev, A, Goody, R.S, Rak, A, Pylypenko, O. | | Deposit date: | 2008-03-31 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism.
J.Biol.Chem., 283, 2008
|
|
8PY3
 
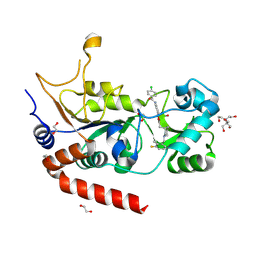 | | Crystal structure of human Sirt2 in complex with a 1,2,4-oxadiazole based inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 4-chloranyl-~{N}-[4-[5-[[(3~{S})-1-[(3-fluoranyl-2-methyl-phenyl)methyl]piperidin-3-yl]methyl]-1,2,4-oxadiazol-3-yl]phenyl]benzamide, ... | | Authors: | Friedrich, F, Colcerasa, A, Einsle, O, Jung, M. | | Deposit date: | 2023-07-24 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure-Activity Studies of 1,2,4-Oxadiazoles for the Inhibition of the NAD + -Dependent Lysine Deacylase Sirtuin 2.
J.Med.Chem., 67, 2024
|
|
3D07
 
 | |
3D43
 
 | | The crystal structure of Sph at 0.8A | | Descriptor: | CALCIUM ION, Sphericase | | Authors: | Almog, O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-04-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (0.8 Å) | | Cite: | The crystal structures of the psychrophilic subtilisin S41 and the mesophilic subtilisin Sph reveal the same calcium-loaded state.
Proteins, 74, 2009
|
|
3D3Z
 
 | | Crystal structure of Actibind a T2 RNase | | Descriptor: | 1,2-ETHANEDIOL, 1-(5-deoxy-beta-L-xylofuranosyl)pyrimidine-2,4(1H,3H)-dione, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gonzalez, A, Almog, O. | | Deposit date: | 2008-05-13 | | Release date: | 2009-06-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structue of Actibind a T2 RNase
To be Published
|
|
3CEH
 
 | | Human liver glycogen phosphorylase (tense state) in complex with the allosteric inhibitor AVE5688 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-[3-(2-Chloro-4,5-difluoro-benzoyl)ureido]-3-trifluoromethoxybenzoic acid, Glycogen phosphorylase, ... | | Authors: | Wendt, K.U, Dreyer, M.K, Anderka, O, Klabunde, T, Loenze, P, Defossa, E, Schmoll, D. | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Thermodynamic characterization of allosteric glycogen phosphorylase inhibitors.
Biochemistry, 47, 2008
|
|
3CEM
 
 | | Human glycogen phosphorylase (tense state) in complex with the allosteric inhibitor AVE9423 | | Descriptor: | 1-(2-carboxyphenyl)-7-chloro-6-[(2-chloro-4,6-difluorophenyl)amino]-4-oxo-1,4-dihydroquinoline-3-carboxylic acid, Glycogen phosphorylase, liver form, ... | | Authors: | Wendt, K.U, Dreyer, M.K, Anderka, O, Klabunde, T, Loenze, P, Defossa, E, Schmoll, D. | | Deposit date: | 2008-02-29 | | Release date: | 2008-05-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Thermodynamic characterization of allosteric glycogen phosphorylase inhibitors.
Biochemistry, 47, 2008
|
|
1WT6
 
 | | Coiled-Coil domain of DMPK | | Descriptor: | Myotonin-protein kinase | | Authors: | Garcia, P, Marino, M, Mayans, O. | | Deposit date: | 2004-11-16 | | Release date: | 2005-11-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Molecular insights into the self-assembly mechanism of dystrophia myotonica kinase
Faseb J., 20, 2006
|
|
3CY2
 
 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and a beta carboline ligand II | | Descriptor: | (4R)-7-chloro-9-methyl-1-oxo-1,2,4,9-tetrahydrospiro[beta-carboline-3,4'-piperidine]-4-carbonitrile, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Huber, K, Bracher, F, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | 7,8-Dichloro-1-oxo-beta-carbolines as a Versatile Scaffold for the Development of Potent and Selective Kinase Inhibitors with Unusual Binding Modes
J.Med.Chem., 55, 2012
|
|
3D09
 
 | |
8UFM
 
 | | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2 | | Descriptor: | ACETATE ION, FORMIC ACID, Papain-like protease nsp3, ... | | Authors: | Minasov, G, Shuvalova, L, Brunzelle, J.S, Rosas-Lemus, M, Kiryukhina, O, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of L516C/Y647C Mutant of SARS-Unique Domain (SUD) from SARS-CoV-2
To Be Published
|
|
8UFL
 
 | | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus | | Descriptor: | CHLORIDE ION, Papain-like protease nsp3, SULFATE ION | | Authors: | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Structural Biology of Infectious Diseases (CSBID) | | Deposit date: | 2023-10-04 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal Structure of SARS-Unique Domain (SUD) of Nsp3 from SARS coronavirus
To Be Published
|
|
3CRF
 
 | |
1DJA
 
 | |
3CLB
 
 | | Structure of bifunctional TcDHFR-TS in complex with TMQ | | Descriptor: | 1,2-ETHANEDIOL, DHFR-TS, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Schormann, N, Senkovich, O, Chattopadhyay, D. | | Deposit date: | 2008-03-18 | | Release date: | 2009-01-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based approach to pharmacophore identification, in silico screening, and three-dimensional quantitative structure-activity relationship studies for inhibitors of Trypanosoma cruzi dihydrofolate reductase function.
Proteins, 73, 2008
|
|
3S95
 
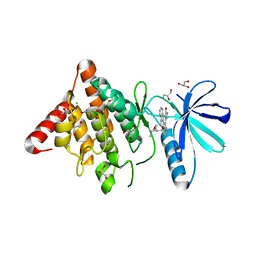 | | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, ... | | Authors: | Beltrami, A, Chaikuad, A, Daga, N, Elkins, J.M, Mahajan, P, Savitsky, P, Vollmar, M, Krojer, T, Muniz, J.R.C, Fedorov, O, Allerston, C.K, Yue, W.W, Gileadi, O, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Bullock, A, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-31 | | Release date: | 2011-07-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the human LIMK1 kinase domain in complex with staurosporine
To be Published
|
|
3D1G
 
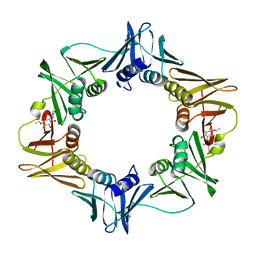 | | Structure of a small molecule inhibitor bound to a DNA sliding clamp | | Descriptor: | DNA polymerase III subunit beta, [(5R)-5-(2,3-dibromo-5-ethoxy-4-hydroxybenzyl)-4-oxo-2-thioxo-1,3-thiazolidin-3-yl]acetic acid | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3CEK
 
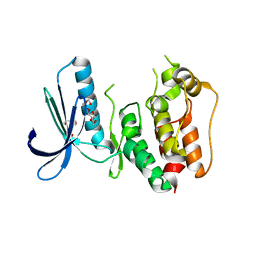 | | Crystal structure of human dual specificity protein kinase (TTK) | | Descriptor: | 2-(2-(2-(2-(2-(2-ETHOXYETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHOXY)ETHANOL, Dual specificity protein kinase TTK | | Authors: | Filippakopoulos, P, Soundararajan, M, Keates, T, Elkins, J.M, King, O, Fedorov, O, Picaud, S.S, Pike, A.C.W, Roos, A, Pilka, E, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-02-29 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Small-molecule kinase inhibitors provide insight into Mps1 cell cycle function.
Nat.Chem.Biol., 6, 2010
|
|
3D0A
 
 | |
3D1E
 
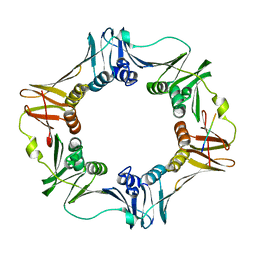 | | Crystal structure of E. coli sliding clamp (beta) bound to a polymerase II peptide | | Descriptor: | DNA polymerase III subunit beta, decamer from polymerase II C-terminal | | Authors: | Georgescu, R.E, Yurieva, O, Seung-Sup, K, Kuriyan, J, Kong, X.-P, O'Donnell, M. | | Deposit date: | 2008-05-05 | | Release date: | 2008-07-29 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a small-molecule inhibitor of a DNA polymerase sliding clamp.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4JNW
 
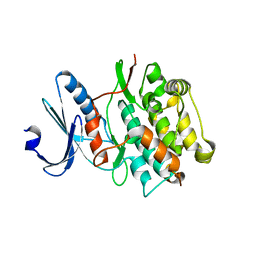 | | Bacterially expressed Titin Kinase | | Descriptor: | GLYCEROL, Titin | | Authors: | Bogomolovas, J, Labeit, S, Mayans, O. | | Deposit date: | 2013-03-16 | | Release date: | 2014-05-14 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Titin kinase is an inactive pseudokinase scaffold that supports MuRF1 recruitment to the sarcomeric M-line.
Open Biol, 4, 2014
|
|
3CY3
 
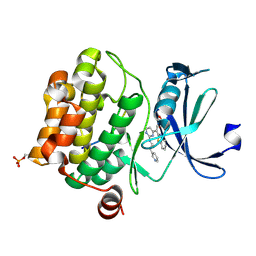 | | Crystal structure of human proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V | | Descriptor: | (2S)-1,3-benzothiazol-2-yl{2-[(2-pyridin-3-ylethyl)amino]pyrimidin-4-yl}ethanenitrile, 1,2-ETHANEDIOL, Pimtide peptide, ... | | Authors: | Filippakopoulos, P, Bullock, A, Fedorov, O, Pike, A.C.W, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-04-25 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Proto-oncogene serine threonine kinase (PIM1) in complex with a consensus peptide and the JNK inhibitor V.
To be Published
|
|
3CPI
 
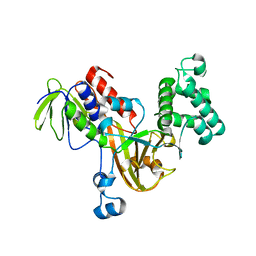 | | Crystal structure of yeast Rab-GDI | | Descriptor: | Rab GDP-dissociation inhibitor | | Authors: | Kravchenko, S, Ignatev, A, Goody, R.S, Rak, A, Pylypenko, O. | | Deposit date: | 2008-03-31 | | Release date: | 2008-05-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A structural model of the GDP dissociation inhibitor rab membrane extraction mechanism.
J.Biol.Chem., 283, 2008
|
|
3D3R
 
 | | Crystal structure of the hydrogenase assembly chaperone HypC/HupF family protein from Shewanella oneidensis MR-1 | | Descriptor: | Hydrogenase assembly chaperone hypC/hupF | | Authors: | Kim, Y, Skarina, T, Onopriyenko, O, Edwards, A.M, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-05-12 | | Release date: | 2008-05-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Hydrogenase Assembly Chaperone HypC/HupF Family Protein from Shewanella oneidensis MR-1.
To be Published
|
|
