4IAF
 
 | |
1MOU
 
 | | Crystal structure of Coral pigment | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
2A4G
 
 | | Hepatitis C Protease NS3-4A serine protease with Ketoamide Inhibitor SCH225724 Bound | | Descriptor: | ({1-[1-CARBAMOYL-PHENYL-METHYL)-CARBAMOYL]-METHYL}-AMINOOXALYL)-BUTYLCARBAMOYL)-3-METHYL-BUTYLCARBAMOYL)-CYCLOHEXYL-METHYL)-CARBAMIC ACID ISOBUTYL ESTER, NS3 protease/helicase, NS4a peptide, ... | | Authors: | Arasappan, A, Njoroge, F.G, Chan, T.Y, Bennett, F, Bogen, S.L, Chen, K, Gu, H, Hong, L, Jao, E, Liu, Y.T, Lovey, R.G, Parekh, T, Pike, R.E, Pinto, P, Santhanam, B, Venkatraman, S, Vaccaro, H, Wang, H, Yang, X, Zhu, Z, Mckittrick, B, Saksena, A.K, Girijavallabhan, V, Pichardo, J, Butkiewicz, N, Ingram, R, Malcolm, B, Prongay, A.J, Yao, N, Marten, B, Madison, V, Kemp, S, Levy, O, Lim-Wilby, M, Tamura, S, Ganguly, A.K. | | Deposit date: | 2005-06-28 | | Release date: | 2006-07-04 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Hepatitis C virus NS3-4a serine protease inhibitors. SAR of P2' moiety with improved potency.
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1M2X
 
 | |
1M8O
 
 | | Platelet integrin alfaIIb-beta3 cytoplasmic domain | | Descriptor: | platelet integrin alfaIIb subunit: cytoplasmic domain, platelet integrin beta3 subunit: cytoplasmic domain | | Authors: | Vinogradova, O, Velyvis, A, Velyviene, A, Hu, B, Haas, T, Plow, E.F, Qin, J. | | Deposit date: | 2002-07-25 | | Release date: | 2002-09-18 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A Structural mechanism of integrin alfaiib beta3
"Inside-out" Activation as regulated by its cytoplasmic face.
Cell(Cambridge,Mass.), 110, 2002
|
|
4IKX
 
 | | Crystal structure of peptide transporter POT (E310Q mutant) | | Descriptor: | Di-tripeptide ABC transporter (Permease), OLEIC ACID, SULFATE ION | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
1M2B
 
 | | Crystal structure at 1.25 Angstroms resolution of the Cys55Ser variant of the thioredoxin-like [2Fe-2S] ferredoxin from Aquifex aeolicus | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, [2Fe-2S] ferredoxin | | Authors: | Yeh, A.P, Ambroggio, X.I, Andrade, S.L.A, Einsle, O, Chatelet, C, Meyer, J, Rees, D.C. | | Deposit date: | 2002-06-22 | | Release date: | 2002-09-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | High-resolution crystal structures
of the wild type and Cys-55-->Ser and
Cys-59-->Ser variants of the thioredoxin-like
[2Fe-2S] ferredoxin from Aquifex aeolicus
J.Biol.Chem., 277, 2002
|
|
1M31
 
 | | Three-Dimensional Solution Structure of Apo-Mts1 | | Descriptor: | Placental calcium-binding protein | | Authors: | Vallely, K.M, Rustandi, R.R, Ellis, K.C, Varlamova, O, Bresnick, A.R, Weber, D.J. | | Deposit date: | 2002-06-26 | | Release date: | 2002-10-30 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of human Mts1 (S100A4) as determined by NMR spectroscopy.
Biochemistry, 41, 2002
|
|
1M4Z
 
 | | Crystal structure of the N-terminal BAH domain of Orc1p | | Descriptor: | MANGANESE (II) ION, ORIGIN RECOGNITION COMPLEX SUBUNIT 1 | | Authors: | Zhang, Z, Hayashi, M.K, Merkel, O, Stillman, B, Xu, R.-M. | | Deposit date: | 2002-07-05 | | Release date: | 2002-09-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and function of the BAH-containing domain of Orc1p in epigenetic silencing.
EMBO J., 21, 2002
|
|
2ABR
 
 | |
3S8Y
 
 | | Bromide soaked structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | BROMIDE ION, Esterase APC40077 | | Authors: | Petit, P, Dong, A, Kagan, O, Savchenko, A, Yakunin, A.F. | | Deposit date: | 2011-05-31 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
1M4W
 
 | | Thermophilic b-1,4-xylanase from Nonomuraea flexuosa | | Descriptor: | ACETATE ION, GLYCEROL, alpha-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Hakulinen, N, Turunen, O, Janis, J, Leisola, M, Rouvinen, J. | | Deposit date: | 2002-07-05 | | Release date: | 2003-07-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Three-dimensional structures of thermophilic beta-1,4-xylanases from Chaetomium thermophilum and Nonomuraea flexuosa. Comparison of twelve xylanases in relation to their thermal stability.
Eur.J.Biochem., 270, 2003
|
|
4IUD
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form with ascorbate - partly reduced state | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Hammer, M, Schmidt, A, Frielingsdorf, S, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
1MOV
 
 | | Crystal structure of Coral protein mutant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
4IUC
 
 | | Crystal structure of an O2-tolerant [NiFe]-hydrogenase from Ralstonia eutropha in its as-isolated form - oxidized state 2 | | Descriptor: | CHLORIDE ION, FE3-S4 CLUSTER, IRON/SULFUR CLUSTER, ... | | Authors: | Frielingsdorf, S, Schmidt, A, Fritsch, J, Lenz, O, Scheerer, P. | | Deposit date: | 2013-01-20 | | Release date: | 2014-04-02 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Reversible [4Fe-3S] cluster morphing in an O2-tolerant [NiFe] hydrogenase.
Nat.Chem.Biol., 10, 2014
|
|
1MJI
 
 | | DETAILED ANALYSIS OF RNA-PROTEIN INTERACTIONS WITHIN THE BACTERIAL RIBOSOMAL PROTEIN L5/5S RRNA COMPLEX | | Descriptor: | 50S ribosomal protein L5, 5S rRNA fragment, MAGNESIUM ION, ... | | Authors: | Perederina, A, Nevskaya, N, Nikonov, O, Nikulin, A, Dumas, P, Yao, M, Tanaka, I, Garber, M, Gongadze, G, Nikonov, S. | | Deposit date: | 2002-08-28 | | Release date: | 2003-01-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detailed analysis of RNA-protein interactions within the bacterial ribosomal
protein L5/5S rRNA complex
RNA, 8, 2002
|
|
1MXD
 
 | | Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
1N1Q
 
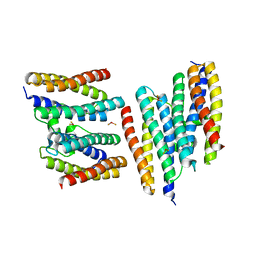 | | Crystal structure of a Dps protein from Bacillus brevis | | Descriptor: | DPS Protein, MU-OXO-DIIRON | | Authors: | Ren, B, Tibbelin, G, Kajino, T, Asami, O, Ladenstein, R. | | Deposit date: | 2002-10-19 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Multi-layered Structure of Dps with a Novel Di-nuclear Ferroxidase Center
J.Mol.Biol., 329, 2003
|
|
1MXF
 
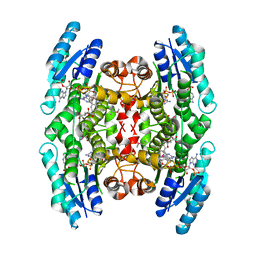 | | Crystal Structure of Inhibitor Complex of Putative Pteridine Reductase 2 (PTR2) from Trypanosoma cruzi | | Descriptor: | METHOTREXATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PTERIDINE REDUCTASE 2 | | Authors: | Schormann, N, Pal, B, Senkovich, O, Carson, M, Howard, A, Smith, C, Delucas, L, Chattopadhyay, D. | | Deposit date: | 2002-10-02 | | Release date: | 2003-10-14 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of Trypanosoma cruzi pteridine reductase 2 in complex with a substrate and an inhibitor.
J.Struct.Biol., 152, 2005
|
|
284D
 
 | | THE BI-LOOP, A NEW GENERAL FOUR-STRANDED DNA MOTIF | | Descriptor: | BARIUM ION, DNA (5'-CD(*P*AP*TP*TP*CP*AP*TP*TP*C)-3'), SODIUM ION | | Authors: | Salisbury, S.A, Wilson, S.E, Powell, H.R, Kennard, O, Lubini, P, Sheldrick, G.M, Escaja, N, Alazzouzi, E, Grandas, A, Pedroso, E. | | Deposit date: | 1996-09-11 | | Release date: | 1997-06-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | The bi-loop, a new general four-stranded DNA motif.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
1MZ6
 
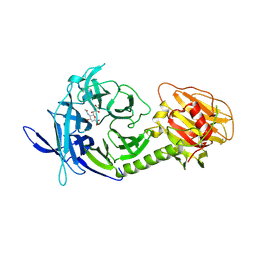 | | Trypanosoma rangeli sialidase in complex with the inhibitor DANA | | Descriptor: | 2-DEOXY-2,3-DEHYDRO-N-ACETYL-NEURAMINIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, sialidase | | Authors: | Buschiazzo, A, Tavares, G.A, Campetella, O, Spinelli, S, Cremona, M.L, Paris, G, Amaya, M.F, Frasch, A.C.C, Alzari, P.M. | | Deposit date: | 2002-10-05 | | Release date: | 2002-10-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of sialyltransferase activity in trypanosomal sialidases
Embo J., 19, 2000
|
|
272D
 
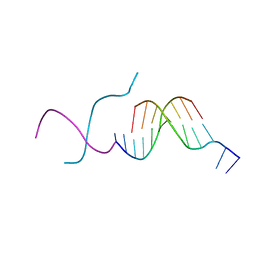 | | PARALLEL AND ANTIPARALLEL (G.GC)2 TRIPLE HELIX FRAGMENTS IN A CRYSTAL STRUCTURE | | Descriptor: | DNA (5'-D(*GP*GP*CP*CP*AP*AP*TP*TP*GP*G)-3') | | Authors: | Vlieghe, D, Van Meervelt, L, Dautant, A, Gallois, B, Precigoux, G, Kennard, O. | | Deposit date: | 1996-07-09 | | Release date: | 1996-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Parallel and antiparallel (G.GC)2 triple helix fragments in a crystal structure.
Science, 273, 1996
|
|
2A9G
 
 | | Structure of C406A arginine deiminase in complex with L-arginine | | Descriptor: | ARGININE, Arginine deiminase | | Authors: | Galkin, A, Lu, X, Dunaway-Mariano, D, Herzberg, O. | | Deposit date: | 2005-07-11 | | Release date: | 2005-08-09 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures Representing the Michaelis Complex and the Thiouronium Reaction Intermediate of Pseudomonas aeruginosa Arginine Deiminase.
J.Biol.Chem., 280, 2005
|
|
2AVI
 
 | |
1MXG
 
 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
