7B0J
 
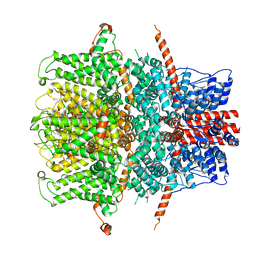 | | TRPC4 in LMNG detergent | | Descriptor: | 2-(HEXADECANOYLOXY)-1-[(PHOSPHONOOXY)METHYL]ETHYL HEXADECANOATE, CALCIUM ION, Transient receptor potential cation channel subfamily c member 4a | | Authors: | Vinayagam, D, Quentin, D, Sistel, O, Merino, F, Stabrin, M, Hofnagel, O, Ledeboer, M.W, Malojcic, G, Raunser, S. | | Deposit date: | 2020-11-20 | | Release date: | 2020-12-09 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural basis of TRPC4 regulation by calmodulin and pharmacological agents.
Elife, 9, 2020
|
|
6AOK
 
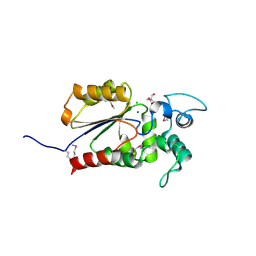 | | Crystal structure of Legionella pneumophila effector Ceg4 with N-terminal TEV protease cleavage sequence | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, Ceg4, ... | | Authors: | Stogios, P.J, Cuff, M.E, Nocek, B, Evdokimova, E, Egorova, O, Yim, V, Di Leo, R, Savchenko, A. | | Deposit date: | 2017-08-16 | | Release date: | 2018-01-10 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | TheLegionella pneumophilaeffector Ceg4 is a phosphotyrosine phosphatase that attenuates activation of eukaryotic MAPK pathways.
J. Biol. Chem., 293, 2018
|
|
5Y50
 
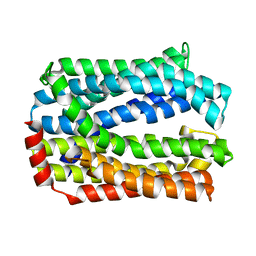 | | Crystal structure of eukaryotic MATE transporter AtDTX14 | | Descriptor: | Protein DETOXIFICATION 14 | | Authors: | Miyauchi, H, Kusakizako, T, Nishizawa, T, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-06 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for xenobiotic extrusion by eukaryotic MATE transporter
Nat Commun, 8, 2017
|
|
5Y78
 
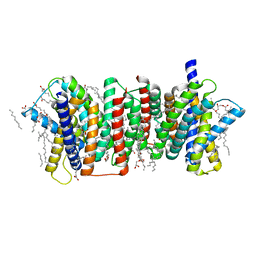 | | Crystal structure of the triose-phosphate/phosphate translocator in complex with inorganic phosphate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, PHOSPHATE ION, Putative hexose phosphate translocator | | Authors: | Lee, Y, Nishizawa, T, Takemoto, M, Kumazaki, K, Yamashita, K, Hirata, K, Minoda, A, Nagatoishi, S, Tsumoto, K, Ishitani, R, Nureki, O. | | Deposit date: | 2017-08-16 | | Release date: | 2017-10-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the triose-phosphate/phosphate translocator reveals the basis of substrate specificity
Nat Plants, 3, 2017
|
|
6BAV
 
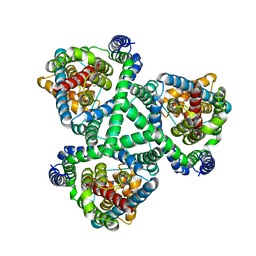 | | Crystal Structure of GltPh R397C in complex with S-Benzyl-L-Cysteine | | Descriptor: | BENZYLCYSTEINE, Glutamate transporter homolog, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-10-16 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6BHR
 
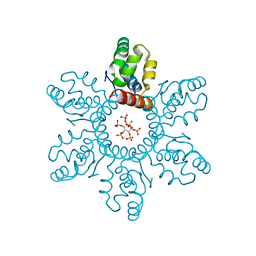 | | HIV-1 immature CTD-SP1 hexamer in complex with IP6 | | Descriptor: | Capsid protein p24,Spacer peptide 1, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
5ZIS
 
 | | Crystal structure of Mn-ProtoporphyrinIX-reconstituted P450BM3 | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, MANGANESE PROTOPORPHYRIN IX | | Authors: | Omura, K, Aiba, Y, Onoda, H, Sugimoto, H, Shoji, O, Watanabe, Y. | | Deposit date: | 2018-03-17 | | Release date: | 2018-08-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Reconstitution of full-length P450BM3 with an artificial metal complex by utilising the transpeptidase Sortase A.
Chem. Commun. (Camb.), 54, 2018
|
|
5ZSU
 
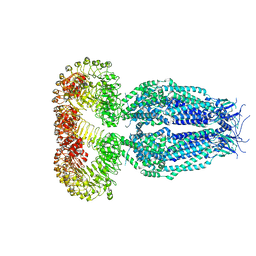 | | Structure of the human homo-hexameric LRRC8A channel at 4.25 Angstroms | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kasuya, G, Nakane, T, Yokoyama, T, Shirouzu, M, Ishitani, R, Nureki, O. | | Deposit date: | 2018-04-29 | | Release date: | 2018-08-15 | | Last modified: | 2018-09-26 | | Method: | ELECTRON MICROSCOPY (4.25 Å) | | Cite: | Cryo-EM structures of the human volume-regulated anion channel LRRC8.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6AEK
 
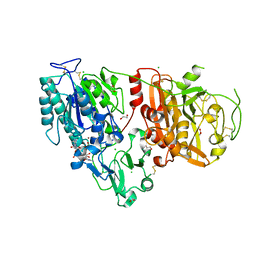 | | Crystal structure of ENPP1 in complex with pApG | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
6BMI
 
 | | Crystal Structure of GltPh R397C in complex with L-Serine | | Descriptor: | Glutamate transporter homolog, SERINE, SODIUM ION | | Authors: | Font, J, Scopelliti, A.J, Vandenberg, R.J, Boudker, O, Ryan, R.M. | | Deposit date: | 2017-11-14 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Structural characterisation reveals insights into substrate recognition by the glutamine transporter ASCT2/SLC1A5.
Nat Commun, 9, 2018
|
|
6BLB
 
 | | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Holliday junction ATP-dependent DNA helicase RuvB, TRIETHYLENE GLYCOL | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | 1.88 Angstrom Resolution Crystal Structure Holliday Junction ATP-dependent DNA Helicase (RuvB) from Pseudomonas aeruginosa in Complex with ADP.
To be Published
|
|
6BHT
 
 | | HIV-1 CA hexamer in complex with IP6, orthorhombic crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.689 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BHS
 
 | | HIV-1 CA hexamer in complex with IP6, hexagonal crystal form | | Descriptor: | Capsid protein p24, INOSITOL HEXAKISPHOSPHATE | | Authors: | Zadrozny, K, Wagner, J.M, Ganser-Pornillos, B.K, Pornillos, O. | | Deposit date: | 2017-10-31 | | Release date: | 2018-08-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Inositol phosphates are assembly co-factors for HIV-1.
Nature, 560, 2018
|
|
6BRH
 
 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
6BRK
 
 | | The SAM domain of mouse SAMHD1 is critical for its activation and regulation | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, Deoxynucleoside triphosphate triphosphohydrolase SAMHD1, MAGNESIUM ION | | Authors: | Buzovetsky, O, Tang, C, Knecht, K.M, Antonucci, J.M, Wu, L, Ji, X, Xiong, Y. | | Deposit date: | 2017-11-30 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | The SAM domain of mouse SAMHD1 is critical for its activation and regulation.
Nat Commun, 9, 2018
|
|
6AI6
 
 | | Crystal structure of SpCas9-NG | | Descriptor: | 1,2-ETHANEDIOL, CRISPR-associated endonuclease Cas9/Csn1, DNA (28-MER), ... | | Authors: | Nishimasu, H, Hirano, S, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-21 | | Release date: | 2018-10-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Engineered CRISPR-Cas9 nuclease with expanded targeting space
Science, 361, 2018
|
|
6AEL
 
 | | Crystal structure of ENPP1 in complex with 3'3'-cGAMP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, ... | | Authors: | Kato, K, Nishimasu, H, Hirano, S, Hirano, H, Ishitani, R, Nureki, O. | | Deposit date: | 2018-08-05 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into cGAMP degradation by Ecto-nucleotide pyrophosphatase phosphodiesterase 1.
Nat Commun, 9, 2018
|
|
6BQ9
 
 | | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida | | Descriptor: | CHLORIDE ION, DNA topoisomerase 4 subunit A, SODIUM ION | | Authors: | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-11-27 | | Release date: | 2017-12-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | 2.55 Angstrom Resolution Crystal Structure of N-terminal Fragment (residues 1-493) of DNA Topoisomerase IV Subunit A from Pseudomonas putida.
To Be Published
|
|
6BRP
 
 | | F-box protein form 2 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
6C4V
 
 | | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis | | Descriptor: | Polyketide synthase Pks13, ZINC ION | | Authors: | Minasov, G, Brunzelle, J.S, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-01-12 | | Release date: | 2018-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1.9 Angstrom Resolution Crystal Structure of Acyl Carrier Protein Domain (residues 1350-1461) of Polyketide Synthase Pks13 from Mycobacterium tuberculosis.
To be Published
|
|
3BQR
 
 | | Crystal structure of human death associated protein kinase 3 (DAPK3) in complex with an imidazo-pyridazine ligand | | Descriptor: | 4-(6-{[(1R)-1-(hydroxymethyl)propyl]amino}imidazo[1,2-b]pyridazin-3-yl)benzoic acid, Death-associated protein kinase 3, GLYCEROL, ... | | Authors: | Filippakopoulos, P, Rellos, P, Fedorov, O, Niesen, F, Pike, A.C.W, Pilka, E.S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-12-20 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of Human Death Associated Protein Kinase 3 (DAPK3) in Complex with an Imidazo-Pyridazine Ligand.
To be Published
|
|
3BTF
 
 | | THE CRYSTAL STRUCTURES OF THE COMPLEXES BETWEEN BOVINE BETA-TRYPSIN AND TEN P1 VARIANTS OF BPTI. | | Descriptor: | CALCIUM ION, PROTEIN (PANCREATIC TRYPSIN INHIBITOR), PROTEIN (TRYPSIN), ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-10 | | Release date: | 2000-03-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3BWU
 
 | | Crystal structure of the ternary complex of FimD (N-Terminal Domain, FimDN) with FimC and the N-terminally truncated pilus subunit FimF (FimFt) | | Descriptor: | 1,2-ETHANEDIOL, Chaperone protein fimC, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Eidam, O, Grutter, M.G, Capitani, G. | | Deposit date: | 2008-01-10 | | Release date: | 2008-03-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of the ternary FimC-FimF(t)-FimD(N) complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD
Febs Lett., 582, 2008
|
|
3BTE
 
 | | The Crystal Structures of the Complexes Between Bovine Beta-Trypsin and Ten P1 Variants of BPTI. | | Descriptor: | CALCIUM ION, PANCREATIC TRYPSIN INHIBITOR, SULFATE ION, ... | | Authors: | Helland, R, Otlewski, J, Sundheim, O, Dadlez, M, Smalas, A.O. | | Deposit date: | 1999-03-11 | | Release date: | 2000-03-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structures of the complexes between bovine beta-trypsin and ten P1 variants of BPTI.
J.Mol.Biol., 287, 1999
|
|
3C6V
 
 | | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus | | Descriptor: | CHLORIDE ION, Probable tautomerase/dehalogenase AU4130, SODIUM ION, ... | | Authors: | Singer, A.U, Binkowski, T.A, Skarina, T, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-02-05 | | Release date: | 2008-02-19 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of AU4130/APC7354, a probable enzyme from the thermophilic fungus Aspergillus fumigatus.
To be Published
|
|
