6HA8
 
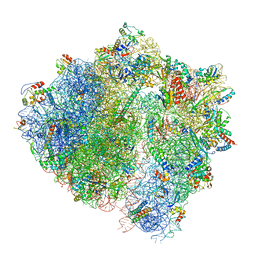 | | Cryo-EM structure of the ABCF protein VmlR bound to the Bacillus subtilis ribosome | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Crowe-McAuliffe, C, Graf, M, Huter, P, Abdelshahid, M, Novacek, J, Wilson, D.N. | | Deposit date: | 2018-08-07 | | Release date: | 2018-08-29 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis for antibiotic resistance mediated by theBacillus subtilisABCF ATPase VmlR.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6HXX
 
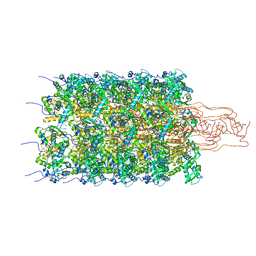 | | Potato virus Y | | Descriptor: | Coat protein, RNA (5'-R(P*UP*UP*UP*UP*U)-3') | | Authors: | Podobnik, M, Kezar, A, Novacek, J, Polak, M. | | Deposit date: | 2018-10-18 | | Release date: | 2019-08-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for the multitasking nature of the potato virus Y coat protein.
Sci Adv, 5, 2019
|
|
5MV6
 
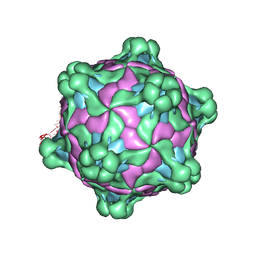 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MUP
 
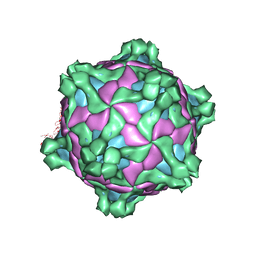 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MV5
 
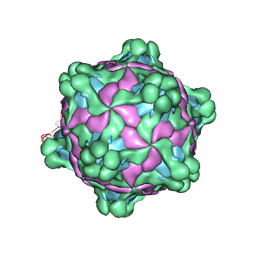 | | Structure of deformed wing virus, a honeybee pathogen | | Descriptor: | URIDINE-5'-MONOPHOSPHATE, VP1, VP2, ... | | Authors: | Skubnik, K, Novacek, J, Fuzik, T, Pridal, A, Paxton, R, Plevka, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of deformed wing virus, a major honey bee pathogen.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5MLC
 
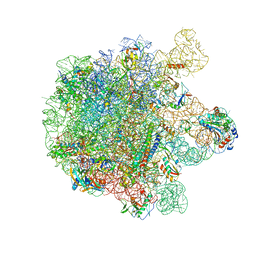 | | Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions | | Descriptor: | 23S ribosomal RNA, chloroplastic, 4.8S ribosomal RNA, ... | | Authors: | Graf, M, Arenz, S, Huter, P, Doenhoefer, A, Novacek, J, Wilson, D.N. | | Deposit date: | 2016-12-06 | | Release date: | 2016-12-28 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structure of the spinach chloroplast ribosome reveals the location of plastid-specific ribosomal proteins and extensions.
Nucleic Acids Res., 45, 2017
|
|
8OOH
 
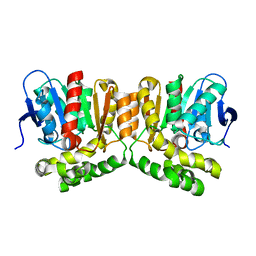 | |
8OKA
 
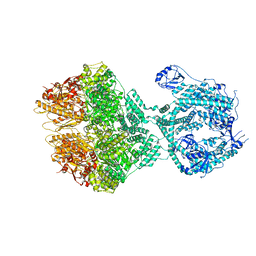 | | Human Mitochondrial Lon Y394F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-28 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OM7
 
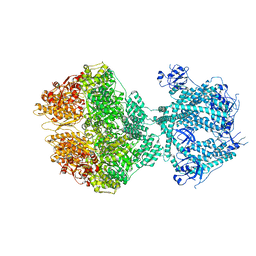 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-31 | | Release date: | 2024-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.74 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OJL
 
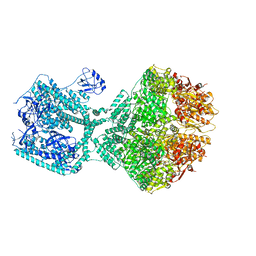 | | Human Mitochondrial Lon Y394E Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OVG
 
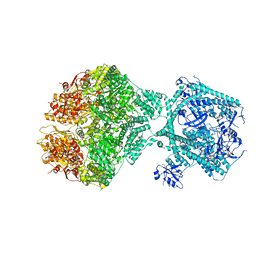 | | Human Mitochondrial Lon Y186E Mutant ADP Bound | | Descriptor: | Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.47 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8OVF
 
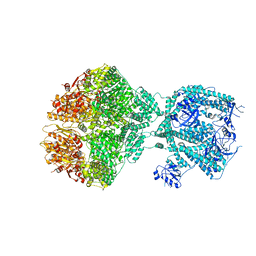 | | Human Mitochondrial Lon Y186F Mutant ADP Bound | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease homolog, mitochondrial | | Authors: | Kereiche, S, Bauer, J.A, Matyas, P, Novacek, J, Kutejova, E. | | Deposit date: | 2023-04-26 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (7.23 Å) | | Cite: | Polyphosphate and tyrosine phosphorylation in the N-terminal domain of the human mitochondrial Lon protease disrupts its functions.
Sci Rep, 14, 2024
|
|
8PUV
 
 | | Trimeric prM/E spike of Tick-borne encephalitis virus immature particle | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope protein E, Protein prM | | Authors: | Fuzik, T, Plevka, P, Smerdova, L, Novacek, J. | | Deposit date: | 2023-07-17 | | Release date: | 2024-07-17 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | The structure of immature tick-borne encephalitis virus supports the collapse model of flavivirus maturation.
Sci Adv, 10, 2024
|
|
8Q2H
 
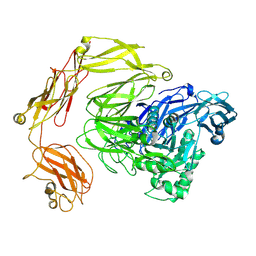 | | beta-galactosidase from Bacillus circulans | | Descriptor: | beta-galactosidase | | Authors: | Kascakova, B, Hovorkova, M, Petraskova, L, Novacek, J, Pinkas, D, Gardian, Z, Kren, V, Bojarova, P, Kuta Smatanova, I. | | Deposit date: | 2023-08-02 | | Release date: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The variable structural flexibility of the Bacillus circulans beta-galactosidase isoforms determines their unique functionalities.
Structure, 2024
|
|
8CBQ
 
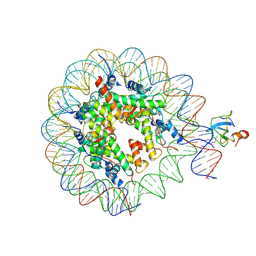 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
8CBN
 
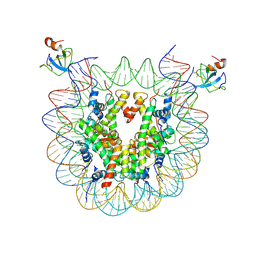 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
7YZ4
 
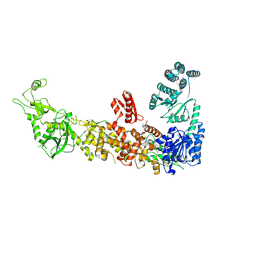 | | Mouse endoribonuclease Dicer (composite structure) | | Descriptor: | Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7YYM
 
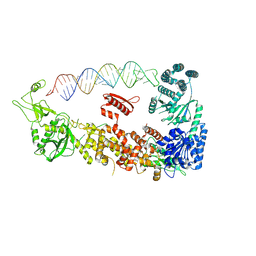 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7YYN
 
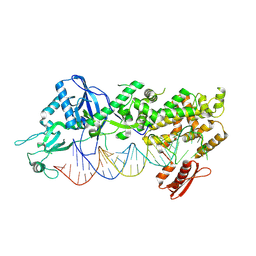 | | Mammalian Dicer in the dicing state with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Isoform 2 of Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-02-18 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (6.21 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7ZPJ
 
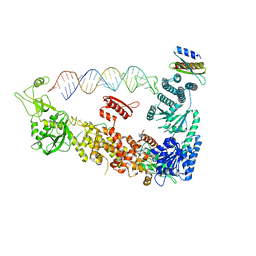 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 isoform 1 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7ZPK
 
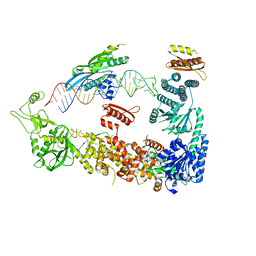 | | Mammalian Dicer in the "pre-dicing state" with pre-miR-15a substrate and TARBP2 subunit | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer, RISC-loading complex subunit TARBP2 | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.81 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
7ZPI
 
 | | Mammalian Dicer in the "dicing state" with pre-miR-15a substrate | | Descriptor: | 59-nt precursor of miR-15a, Endoribonuclease Dicer | | Authors: | Zanova, M, Zapletal, D, Kubicek, K, Stefl, R, Pinkas, M, Novacek, J. | | Deposit date: | 2022-04-27 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Structural and functional basis of mammalian microRNA biogenesis by Dicer.
Mol.Cell, 82, 2022
|
|
8BRJ
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) trimeric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-23 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.08 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
8BNU
 
 | | Escherichia coli anaerobic fatty acid beta oxidation trifunctional enzyme (anEcTFE) tetrameric complex | | Descriptor: | 3-ketoacyl-CoA thiolase FadI, Fatty acid oxidation complex subunit alpha | | Authors: | Sah-Teli, S.K, Pinkas, M, Novacek, J, Venkatesan, R. | | Deposit date: | 2022-11-14 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Structural basis for different membrane-binding properties of E. coli anaerobic and human mitochondrial beta-oxidation trifunctional enzymes.
Structure, 31, 2023
|
|
7Z45
 
 | | Central part (C10) of bacteriophage SU10 capsid | | Descriptor: | Major head protein | | Authors: | Siborova, M, Fuzik, T, Prochazkova, M, Novacek, J, Plevka, P. | | Deposit date: | 2022-03-03 | | Release date: | 2022-08-24 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Tail proteins of phage SU10 reorganize into the nozzle for genome delivery.
Nat Commun, 13, 2022
|
|
