7VPK
 
 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2P state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPI
 
 | | Cryo-EM structure of the human ATP13A2 (E1-ATP state) | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Polyamine-transporting ATPase 13A2 | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPL
 
 | | Cryo-EM structure of the human ATP13A2 (SPM-bound E2Pi state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MAGNESIUM ION, Polyamine-transporting ATPase 13A2, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VPJ
 
 | | Cryo-EM structure of the human ATP13A2 (E1P-ADP state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Tomita, A, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-17 | | Release date: | 2021-12-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Cryo-EM reveals mechanistic insights into lipid-facilitated polyamine export by human ATP13A2.
Mol.Cell, 81, 2021
|
|
7VTI
 
 | | Crystal structure of the Cas13bt3-crRNA binary complex | | Descriptor: | 1,2-ETHANEDIOL, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Nakagawa, R, Takeda, N.S, Tomita, A, Hirano, H, Kusakizako, T, Nishizawa, T, Yamashita, K, Nishimasu, H, Nureki, O. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-24 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure and engineering of the minimal type VI CRISPR-Cas13bt3.
Mol.Cell, 82, 2022
|
|
7V73
 
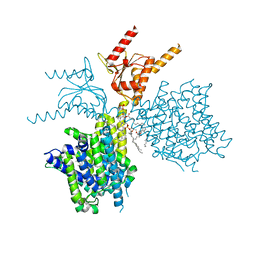 | | Thermostabilized human prestin in complex with chloride | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHLORIDE ION, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V74
 
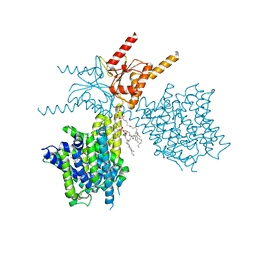 | | Thermostabilized human prestin in complex with sulfate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, CHOLESTEROL, SULFATE ION, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7V75
 
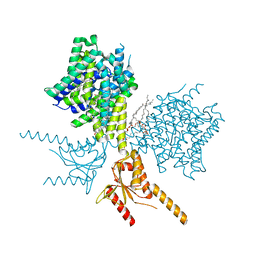 | | Thermostabilized human prestin in complex with salicylate | | Descriptor: | 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine, 2-HYDROXYBENZOIC ACID, CHOLESTEROL, ... | | Authors: | Futamata, H, Fukuda, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-08-21 | | Release date: | 2022-08-31 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Cryo-EM structures of thermostabilized prestin provide mechanistic insights underlying outer hair cell electromotility.
Nat Commun, 13, 2022
|
|
7VR7
 
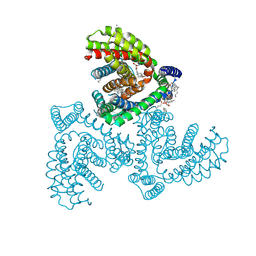 | | Inward-facing structure of human EAAT2 in the WAY213613-bound state | | Descriptor: | (2S)-2-azanyl-4-[[4-[2-bromanyl-4,5-bis(fluoranyl)phenoxy]phenyl]amino]-4-oxidanylidene-butanoic acid, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7E6Z
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 50 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6Y
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E6X
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 4 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E71
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 1 ms structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7E70
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: 250 microsecond structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Archaeal-type opsin 1,Archaeal-type opsin 2, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-02-24 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
7VVL
 
 | | PTH-bound human PTH1R in complex with Gs (class2) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVN
 
 | | PTH-bound human PTH1R in complex with Gs (class4) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVK
 
 | | PTH-bound human PTH1R in complex with Gs (class1) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVM
 
 | | PTH-bound human PTH1R in complex with Gs (class3) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVO
 
 | | PTH-bound human PTH1R in complex with Gs (class5) | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VVJ
 
 | | PTHrP-bound human PTH1R in complex with Gs | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Kobayashi, K, Kusakizako, T, Miyauchi, H, Tomita, A, Kobayashi, K, Shihoya, W, Yamashita, K, Nishizawa, T, Kato, H.E, Nureki, O. | | Deposit date: | 2021-11-06 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Endogenous ligand recognition and structural transition of a human PTH receptor.
Mol.Cell, 82, 2022
|
|
7VR8
 
 | | Inward-facing structure of human EAAT2 in the substrate-free state | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL, ... | | Authors: | Kato, T, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-10-22 | | Release date: | 2022-08-10 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural insights into inhibitory mechanism of human excitatory amino acid transporter EAAT2.
Nat Commun, 13, 2022
|
|
7C86
 
 | | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin: Dark state structure | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, RETINAL, ... | | Authors: | Oda, K, Nomura, T, Nakane, T, Yamashita, K, Inoue, K, Ito, S, Vierock, J, Hirata, K, Maturana, A.D, Katayama, K, Ikuta, T, Ishigami, I, Izume, T, Umeda, R, Eguma, R, Oishi, S, Kasuya, G, Kato, T, Kusakizako, T, Shihoya, W, Shimada, H, Takatsuji, T, Takemoto, M, Taniguchi, R, Tomita, A, Nakamura, R, Fukuda, M, Miyauchi, H, Lee, Y, Nango, E, Tanaka, R, Tanaka, T, Sugahara, M, Kimura, T, Shimamura, T, Fujiwara, T, Yamanaka, Y, Owada, S, Joti, Y, Tono, K, Ishitani, R, Hayashi, S, Kandori, H, Hegemann, P, Iwata, S, Kubo, M, Nishizawa, T, Nureki, O. | | Deposit date: | 2020-05-28 | | Release date: | 2021-04-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Time-resolved serial femtosecond crystallography reveals early structural changes in channelrhodopsin.
Elife, 10, 2021
|
|
6K7H
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1 state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
6K7I
 
 | | Cryo-EM structure of the human P4-type flippase ATP8A1-CDC50 (E1-ATP state class2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHOLESTEROL HEMISUCCINATE, Cell cycle control protein 50A, ... | | Authors: | Hiraizumi, M, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-06-07 | | Release date: | 2019-08-28 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Cryo-EM structures capture the transport cycle of the P4-ATPase flippase.
Science, 365, 2019
|
|
7EK2
 
 | | Cryo-EM structure of VCCN1 in lipid nanodisc | | Descriptor: | Bestrophin-like protein | | Authors: | Hagino, T, Kato, T, Kasuya, G, Kobayashi, K, Kusakizako, T, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2021-04-03 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of thylakoid-located voltage-dependent chloride channel VCCN1.
Nat Commun, 13, 2022
|
|
