4V2U
 
 | | Apo-structure of alpha2,3-sialyltransferase from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-15 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
1Z9A
 
 | | Crystal Structure Of The Asn-309 To Asp Mutant Of Candida Tenuis Xylose Reductase (Akr2B5) Bound To Nad+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Kratzer, R, Leitgeb, S, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2005-04-01 | | Release date: | 2006-01-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Probing the substrate binding site of Candida tenuis xylose reductase (AKR2B5) with site-directed mutagenesis
Biochem.J., 393, 2006
|
|
1LJ8
 
 | | Crystal structure of mannitol dehydrogenase in complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, mannitol dehydrogenase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2002-04-19 | | Release date: | 2002-11-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of Pseudomonas fluorescens mannitol 2-dehydrogenase binary and ternary
complexes. Specificity and catalytic mechanism
J.Biol.Chem., 277, 2002
|
|
8RR2
 
 | | 3-keto-glycoside eliminase/hydratase in komplex with alpha-3-keto-glucose | | Descriptor: | (2~{R},3~{R},5~{S},6~{S})-2-(hydroxymethyl)-3,5,6-tris(oxidanyl)oxan-4-one, 1,2-ETHANEDIOL, 3-keto-disaccharide hydrolase domain-containing protein, ... | | Authors: | Pfeiffer, M, Kastner, K, Oberdorfer, G, Nidetzky, B. | | Deposit date: | 2024-01-22 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8BVK
 
 | | The crystal structure of O-glycoside cleaving beta-eliminase from A. tumefaciens AtOGE | | Descriptor: | MANGANESE (II) ION, Xylose isomerase | | Authors: | Kuhlmann, K, Bitter, J, Pfeiffer, M, Nidetzky, B, Pavkov-Keller, T. | | Deposit date: | 2022-12-04 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enzymatic beta-elimination in natural product O- and C-glycoside deglycosylation.
Nat Commun, 14, 2023
|
|
6Q94
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (S156D) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
4YL2
 
 | | Aerococcus viridans L-lactate oxidase Y191F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Lactate oxidase, PYRUVIC ACID | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2015-03-04 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Speeding up the product release: a second-sphere contribution from Tyr191 to the reactivity of l-lactate oxidase revealed in crystallographic and kinetic studies of site-directed variants.
Febs J., 282, 2015
|
|
3KHU
 
 | | Crystal structure of human UDP-glucose dehydrogenase Glu161Gln, in complex with thiohemiacetal intermediate | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, UDP-glucose 6-dehydrogenase, ... | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Guo, K, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-30 | | Release date: | 2009-11-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and Kinetic Evidence That Catalytic Reaction of Human UDP-glucose 6-Dehydrogenase Involves Covalent Thiohemiacetal and Thioester Enzyme Intermediates.
J.Biol.Chem., 287, 2012
|
|
6RMR
 
 | |
2C4M
 
 | |
5EBU
 
 | | Aerococcus viridans L-lactate oxidase Y215F mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, L-lactate oxidase, PYRUVIC ACID | | Authors: | Rainer, D, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2015-10-19 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational flexibility related to enzyme activity: evidence for a dynamic active-site gatekeeper function of Tyr(215) in Aerococcus viridans lactate oxidase.
Sci Rep, 6, 2016
|
|
6GPJ
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4F-Man | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
3ITK
 
 | | Crystal structure of human UDP-glucose dehydrogenase Thr131Ala, apo form. | | Descriptor: | 1,2-ETHANEDIOL, TETRAETHYLENE GLYCOL, UDP-glucose 6-dehydrogenase | | Authors: | Chaikuad, A, Egger, S, Yue, W.W, Sethi, R, Filippakopoulos, P, Muniz, J.R.C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A.M, Kavanagh, K.L, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-08-28 | | Release date: | 2009-09-15 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure and mechanism of human UDP-glucose 6-dehydrogenase.
J.Biol.Chem., 286, 2011
|
|
8RO4
 
 | | The crystal structure of 2-hydroxy-3-keto-glucal hydratase AtHYD from A. tumefaciens | | Descriptor: | 2-hydroxy-3-keto-glucal hydratase, MANGANESE (II) ION | | Authors: | Grininger, C, Bitter, J, Pfeiffer, M, Nidetzky, B, Pavkov-Keller, T. | | Deposit date: | 2024-01-11 | | Release date: | 2024-08-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Enzyme Machinery for Bacterial Glucoside Metabolism through a Conserved Non-hydrolytic Pathway.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1SM9
 
 | | Crystal Structure Of An Engineered K274RN276D Double Mutant of Xylose Reductase From Candida Tenuis Optimized To Utilize NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, xylose reductase | | Authors: | Petschacher, B, Leitgeb, S, Kavanagh, K.L, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-03-08 | | Release date: | 2004-12-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The coenzyme specificity of Candida tenuis xylose reductase (AKR2B5) explored by site-directed mutagenesis and X-ray crystallography.
Biochem.J., 385, 2005
|
|
4UD8
 
 | | AtBBE15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3,6,9,12,15,18,21,24-OCTAOXAHEXACOSAN-1-OL, ... | | Authors: | Daniel, B, Steiner, B, Pavkov-Keller, T, Dordic, A, Gutmann, A, Sensen, C.W, Nidetzky, B, van der Graaff, E, Wallner, S, Gruber, K, Macheroux, P. | | Deposit date: | 2014-12-09 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Oxidation of Monolignols by Members of the Berberine Bridge Enzyme Family Suggests a Role in Cell Wall Metabolism.
J.Biol.Chem., 290, 2015
|
|
1YE4
 
 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
FEBS Lett., 579, 2005
|
|
1YE6
 
 | | Crystal structure of the Lys-274 to Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NADP+ | | Descriptor: | NAD(P)H-dependent D-xylose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Leitgeb, S, Petschacher, B, Wilson, D.K, Nidetzky, B. | | Deposit date: | 2004-12-28 | | Release date: | 2005-02-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Fine tuning of coenzyme specificity in family 2 aldo-keto reductases revealed by crystal structures of the Lys-274-->Arg mutant of Candida tenuis xylose reductase (AKR2B5) bound to NAD(+) and NADP(+).
Febs Lett., 579, 2005
|
|
6GPK
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase (E157Q) in complex with GDP-Man | | Descriptor: | 1,2-ETHANEDIOL, GDP-mannose 4,6 dehydratase, GLYCEROL, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
6GPL
 
 | | Crystal structure of human GDP-D-mannose 4,6-dehydratase in complex with GDP-4k6d-Man | | Descriptor: | 1,2-ETHANEDIOL, BICINE, GDP-mannose 4,6 dehydratase, ... | | Authors: | Pfeiffer, M, Krojer, T, Johansson, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Nidetzky, B, Oppermann, U, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-06-06 | | Release date: | 2018-07-18 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A Parsimonious Mechanism of Sugar Dehydration by Human GDP-Mannose-4,6-dehydratase.
Acs Catalysis, 9, 2019
|
|
4V3B
 
 | | The structure of alpha2,3-sialyltransferase variant 1 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V3C
 
 | | The structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis in complex with the donor product CMP | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
4V39
 
 | | Apo-structure of alpha2,3-sialyltransferase variant 2 from Pasteurella dagmatis | | Descriptor: | SIALYLTRANSFERASE | | Authors: | Pavkov-Keller, T, Schmoelzer, K, Czabany, T, Luley-Goedl, C, Ribitsch, D, Schwab, H, Nidetzky, B, Gruber, K. | | Deposit date: | 2014-10-17 | | Release date: | 2015-04-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complete Switch from Alpha2,3- to Alpha2,6-Regioselectivity in Pasteurella Dagmatis Beta-D-Galactoside Sialyltransferase by Active-Site Redesign
Chem.Commun.(Camb.), 51, 2015
|
|
1K8C
 
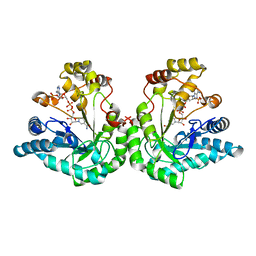 | | Crystal structure of dimeric xylose reductase in complex with NADP(H) | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, xylose reductase | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-10-23 | | Release date: | 2002-07-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
1JEZ
 
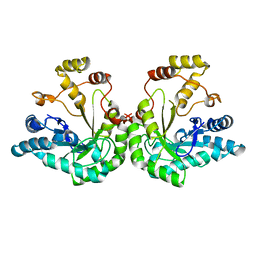 | | THE STRUCTURE OF XYLOSE REDUCTASE, A DIMERIC ALDO-KETO REDUCTASE FROM CANDIDA TENUIS | | Descriptor: | XYLOSE REDUCTASE | | Authors: | Kavanagh, K.L, Klimacek, M, Nidetzky, B, Wilson, D.K. | | Deposit date: | 2001-06-19 | | Release date: | 2002-07-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of apo and holo forms of xylose reductase, a dimeric aldo-keto reductase from Candida tenuis.
Biochemistry, 41, 2002
|
|
