7PU1
 
 | | High resolution X-ray structure of Thermoascus aurantiacus LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Banerjee, S, Frandsen, K.E.H, Singh, R.K, Bjerrum, M.J, Lo Leggio, L. | | Deposit date: | 2021-09-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Protonation State of an Important Histidine from High Resolution Structures of Lytic Polysaccharide Monooxygenases.
Biomolecules, 12, 2022
|
|
7PXW
 
 | | LPMO, expressed in E.coli, in complex with Cellotetraose | | Descriptor: | Auxiliary activity 9, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Banerjee, S, Muderspach, S.J, Tandrup, T, Ipsen, J, Rollan, C.H, Norholm, M, Johansen, K.S, Lo Leggio, L. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Changes in active-site geometry on X-ray photoreduction of a lytic polysaccharide monooxygenase active-site copper and saccharide binding.
Iucrj, 9, 2022
|
|
6FC1
 
 | |
6FC0
 
 | | Crystal structure of the eIF4E-eIF4G complex from Chaetomium thermophilum | | Descriptor: | Eukaryotic translation initiation factor 4E-like protein, Eukaryotic translation initiation factor 4G | | Authors: | Gruener, S, Valkov, E. | | Deposit date: | 2017-12-20 | | Release date: | 2018-06-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.293 Å) | | Cite: | Structural motifs in eIF4G and 4E-BPs modulate their binding to eIF4E to regulate translation initiation in yeast.
Nucleic Acids Res., 46, 2018
|
|
6GDC
 
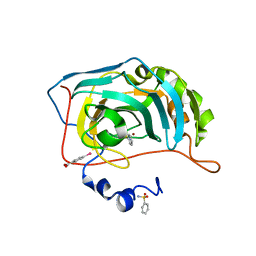 | | Human Carbonic Anhydrase II in complex with Benzenesulfonamide | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, MERCURY (II) ION, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-04-23 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.079 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6GM9
 
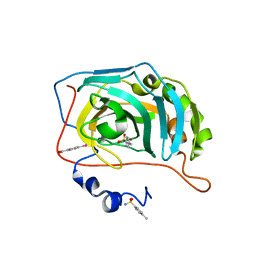 | | Human Carbonic Anhydrase II in complex with 4-Methylbenzenesulfonamide | | Descriptor: | 4-methylbenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-05-24 | | Release date: | 2019-06-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.089 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6I0W
 
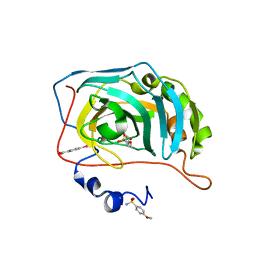 | | Human Carbonic Anhydrase II in complex with 4-Methoxybenzenesulfonamide | | Descriptor: | 4-methoxybenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-10-26 | | Release date: | 2019-11-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.04 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6HR3
 
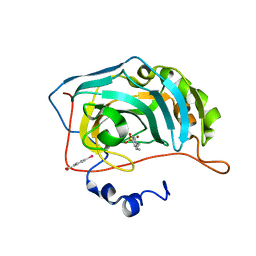 | | Human Carbonic Anhydrase II in complex with 4-Propylbenzenesulfonamide | | Descriptor: | 4-propylbenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.02 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6HXD
 
 | | Human Carbonic Anhydrase II in complex with 4-Butylbenzenesulfonamide | | Descriptor: | 4-butylbenzenesulfonamide, Carbonic anhydrase 2, MERCURIBENZOIC ACID, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-10-17 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
6HQX
 
 | | Human Carbonic Anhydrase II in complex with 4-Ethylbenzenesulfonamide | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-ethylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Heine, A, Klebe, G. | | Deposit date: | 2018-09-25 | | Release date: | 2019-10-09 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.097 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
5E3V
 
 | |
5EMY
 
 | | Human Pancreatic Alpha-Amylase in complex with the mechanism based inactivator glucosyl epi-cyclophellitol | | Descriptor: | (1R,2R,3S,5R,6S)-2,3,5-trihydroxy-6-(hydroxymethyl)cyclohexyl alpha-D-glucopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-11-06 | | Release date: | 2016-07-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.231 Å) | | Cite: | Glucosyl epi-cyclophellitol allows mechanism-based inactivation and structural analysis of human pancreatic alpha-amylase.
Febs Lett., 590, 2016
|
|
5E0F
 
 | | Human pancreatic alpha-amylase in complex with mini-montbretin A | | Descriptor: | 5,7-dihydroxy-4-oxo-2-(3,4,5-trihydroxyphenyl)-4H-chromen-3-yl 6-deoxy-2-O-{6-O-[(2E)-3-(3,4-dihydroxyphenyl)prop-2-enoyl]-beta-D-glucopyranosyl}-alpha-L-mannopyranoside, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Caner, S, Brayer, G.D. | | Deposit date: | 2015-09-28 | | Release date: | 2016-09-07 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Human pancreatic alpha-amylase in complex with mini-montbretin A
To Be Published
|
|
3PVX
 
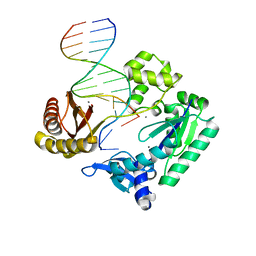 | |
3PW0
 
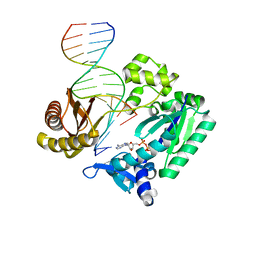 | |
3PW7
 
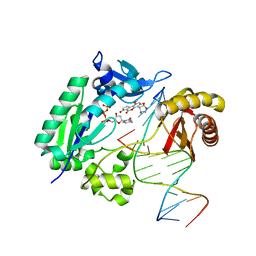 | |
4JV1
 
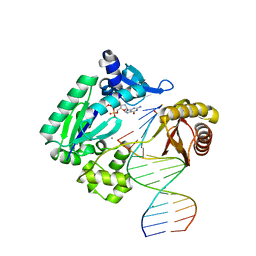 | | Ternary complex of gamma-OHPDG adduct modified dna with dna (-1 primer) polymerase iv and incoming dgtp | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
4JUZ
 
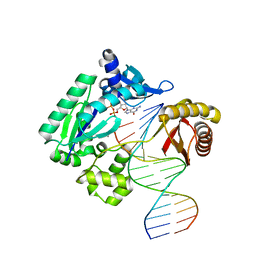 | | Ternary complex of gamma-OHPDG adduct modified dna (zero primer) with dna polymerase iv and incoming dgtp | | Descriptor: | 2'-DEOXYGUANOSINE-5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
3PW5
 
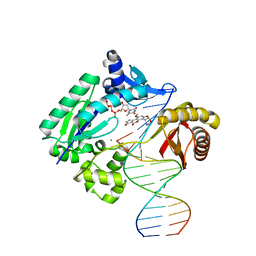 | |
5I7F
 
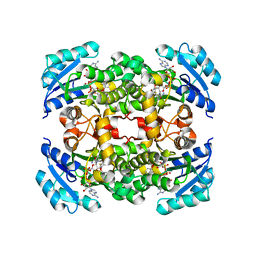 | |
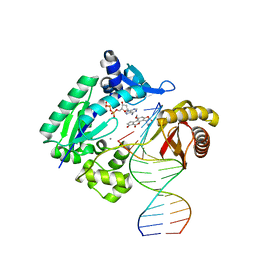
3PW2
 
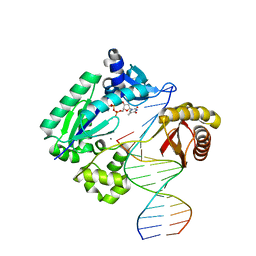 | |
3PW4
 
 | |
4JV2
 
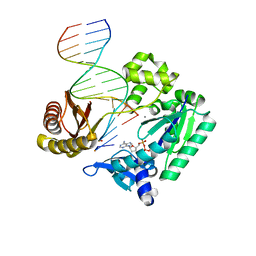 | | Ternary complex of gamma-OHPDG adduct modified dna with dna (-1 primer) polymerase iv and incoming datp | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
4JV0
 
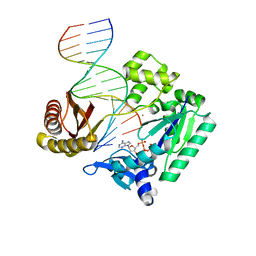 | | Ring-Opening of the -OH-PdG Adduct in Ternary Complexes with the Sulfolobus solfataricus DNA polymerase Dpo4 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, CALCIUM ION, DNA (5'-D(*GP*GP*GP*GP*GP*AP*AP*GP*GP*AP*TP*TP*CP*C)-3'), ... | | Authors: | Banerjee, S, Shanmugam, G, Stone, M.P. | | Deposit date: | 2013-03-25 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Ring-Opening of the gamma-OH-PdG Adduct Promotes Error-Free Bypass by the Sulfolobus solfataricus DNA Polymerase Dpo4.
Chem.Res.Toxicol., 26, 2013
|
|
5UGS
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT501 | | Descriptor: | 5-[(4-cyclopropyl-1,2,3-triazol-1-yl)methyl]-2-(2-methylphenoxy)phenol, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
