5B3D
 
 | | Structure of a flagellar type III secretion chaperone, FlgN | | Descriptor: | Flagella synthesis protein FlgN | | Authors: | Nakanishi, Y, Kinoshita, M, Namba, K, Minamino, T, Imada, K. | | Deposit date: | 2016-02-15 | | Release date: | 2016-06-01 | | Last modified: | 2020-02-26 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Rearrangements of alpha-helical structures of FlgN chaperone control the binding affinity for its cognate substrates during flagellar type III export
Mol.Microbiol., 101, 2016
|
|
2M1X
 
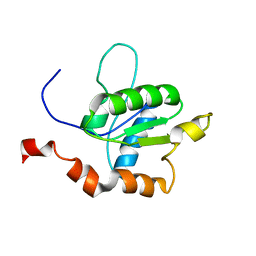 | | TICAM-1 TIR domain structure | | Descriptor: | TIR domain-containing adapter molecule 1 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8HLB
 
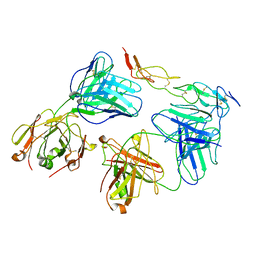 | | Cryo-EM structure of biparatopic antibody Bp109-92 in complex with TNFR2 | | Descriptor: | TR109 heavy chain, TR109 light chain, TR92 heavy chain, ... | | Authors: | Akiba, H, Fujita, J, Ise, T, Nishiyama, K, Miyata, T, Kato, T, Namba, K, Ohno, H, Kamada, H, Nagata, S, Tsumoto, K. | | Deposit date: | 2022-11-29 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (3.63 Å) | | Cite: | Development of a 1:1-binding biparatopic anti-TNFR2 antagonist by reducing signaling activity through epitope selection.
Commun Biol, 6, 2023
|
|
3J82
 
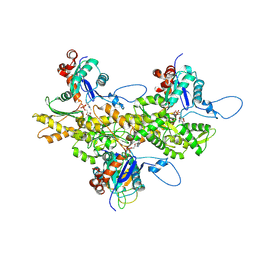 | | Electron cryo-microscopy of DNGR-1 in complex with F-actin | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, cytoplasmic 1, ... | | Authors: | Hanc, P, Fujii, T, Yamada, Y, Huotari, J, Schulz, O, Ahrens, S, Kjaer, S, Way, M, Namba, K, Reis e Sousa, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-05-20 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.7 Å) | | Cite: | Structure of the Complex of F-Actin and DNGR-1, a C-Type Lectin Receptor Involved in Dendritic Cell Cross-Presentation of Dead Cell-Associated Antigens.
Immunity, 42, 2015
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
7VQ0
 
 | | Cryo-EM structure of the SARS-CoV-2 spike protein (2-up RBD) bound to neutralizing nanobodies P86 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
3MFP
 
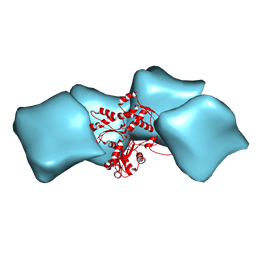 | | Atomic model of F-actin based on a 6.6 angstrom resolution cryoEM map | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Actin, alpha skeletal muscle | | Authors: | Fujii, T, Iwane, A.H, Yanagida, T, Namba, K. | | Deposit date: | 2010-04-03 | | Release date: | 2010-09-29 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Direct visualization of secondary structures of F-actin by electron cryomicroscopy
Nature, 467, 2010
|
|
2M1W
 
 | | TICAM-2 TIR domain | | Descriptor: | TIR domain-containing adapter molecule 2 | | Authors: | Enokizono, Y, Kumeta, H, Funami, K, Horiuchi, M, Sarmiento, J, Yamashita, K, Standley, D.M, Matsumoto, M, Seya, T, Inagaki, F. | | Deposit date: | 2012-12-07 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structures and interface mapping of the TIR domain-containing adaptor molecules involved in interferon signaling.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
8GY2
 
 | | Cryo-EM Structure of Membrane-Bound Alcohol Dehydrogenase from Gluconobacter oxydans | | Descriptor: | Alcohol dehydrogenase (quinone), cytochrome c subunit, dehydrogenase subunit, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
8GY3
 
 | | Cryo-EM Structure of Membrane-Bound Aldehyde Dehydrogenase from Gluconobacter oxydans | | Descriptor: | (MOLYBDOPTERIN-CYTOSINE DINUCLEOTIDE-S,S)-DIOXO-AQUA-MOLYBDENUM(V), Cytochrome c subunit of aldehyde dehydrogenase, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Adachi, T, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2022-09-21 | | Release date: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Experimental and Theoretical Insights into Bienzymatic Cascade for Mediatorless Bioelectrochemical Ethanol Oxidation with Alcohol and Aldehyde Dehydrogenases
Acs Catalysis, 13, 2023
|
|
6JZT
 
 | |
8JEK
 
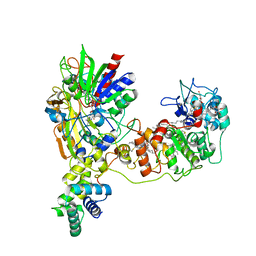 | | Cryo-EM Structure of K-ferricyanide Oxidized Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
8JEJ
 
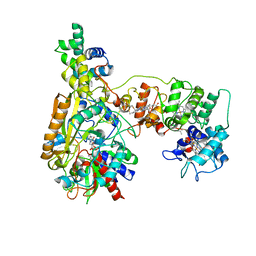 | | Cryo-EM Structure of Na-dithionite Reduced Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Suzuki, Y, Miyata, T, Makino, F, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-05-16 | | Release date: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Essential Insight of Direct Electron Transfer-Type Bioelectrocatalysis by Membrane-Bound d-Fructose Dehydrogenase with Structural Bioelectrochemistry.
Acs Catalysis, 13, 2023
|
|
2MGS
 
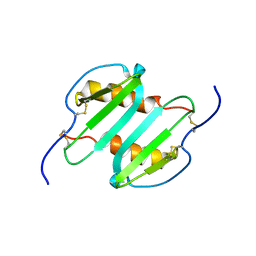 | | Solution structure of CXCL5 | | Descriptor: | C-X-C motif chemokine 5 | | Authors: | Sepuru, K, Rajarathnam, K. | | Deposit date: | 2013-11-04 | | Release date: | 2014-04-02 | | Last modified: | 2016-04-27 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of CXCL5 - A Novel Chemokine and Adipokine Implicated in Inflammation and Obesity.
Plos One, 9, 2014
|
|
8W8M
 
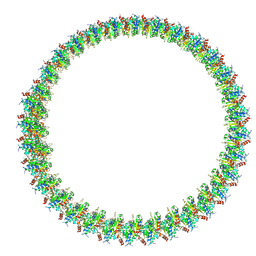 | | Cryo-EM structure of helical filament of MyD88 TIR | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Kasai, K, Imamura, K, Narita, A, Makino, F, Miyata, T, Kato, T, Namba, K, Onishi, H, Tochio, H. | | Deposit date: | 2023-09-04 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Signaling adopter protein in a self-assembled form
To Be Published
|
|
6JZR
 
 | |
8H1O
 
 | | Cryo-EM structure of KpFtsZ-monobody double helical tube | | Descriptor: | Cell division protein FtsZ, GUANOSINE-5'-DIPHOSPHATE, Mb(Ec/KpFtsZ_S1) | | Authors: | Fujita, J, Amesaka, H, Yoshizawa, T, Kuroda, N, Kamimura, N, Hara, M, Inoue, T, Namba, K, Tanaka, S, Matsumura, H. | | Deposit date: | 2022-10-03 | | Release date: | 2023-08-02 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structures of a FtsZ single protofilament and a double-helical tube in complex with a monobody.
Nat Commun, 14, 2023
|
|
7VW6
 
 | | Cryo-EM Structure of Formate Dehydrogenase 1 from Methylorubrum extorquens AM1 | | Descriptor: | 2-AMINO-5,6-DIMERCAPTO-7-METHYL-3,7,8A,9-TETRAHYDRO-8-OXA-1,3,9,10-TETRAAZA-ANTHRACEN-4-ONE GUANOSINE DINUCLEOTIDE, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Yoshikawa, T, Makino, F, Miyata, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2021-11-09 | | Release date: | 2022-06-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.19 Å) | | Cite: | Multiple electron transfer pathways of tungsten-containing formate dehydrogenase in direct electron transfer-type bioelectrocatalysis.
Chem.Commun.(Camb.), 58, 2022
|
|
2BGZ
 
 | | ATOMIC MODEL OF THE BACTERIAL FLAGELLAR BASED ON DOCKING AN X-RAY DERIVED HOOK STRUCTURE INTO AN EM MAP. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, Derosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8K6J
 
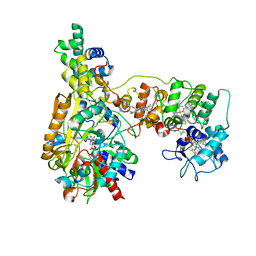 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-H1147A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (H1147A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8K6K
 
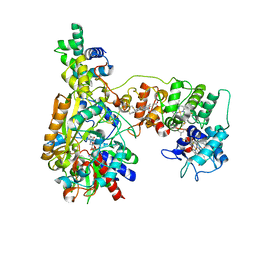 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase (N1146A) large subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-07-25 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
2BGY
 
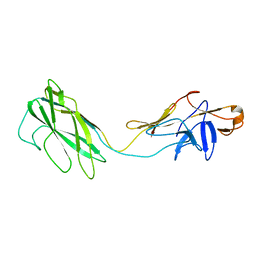 | | Fit of the x-ray structure of the baterial flagellar hook fragment flge31 into an EM map from the hook of Caulobacter crescentus. | | Descriptor: | FLAGELLAR HOOK PROTEIN FLGE | | Authors: | Shaikh, T.R, Thomas, D.R, Chen, J.Z, Samatey, F.A, Matsunami, H, Imada, K, Namba, K, DeRosier, D.J. | | Deposit date: | 2005-01-06 | | Release date: | 2005-01-27 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (9 Å) | | Cite: | A Partial Atomic Structure for the Flagellar Hook of Salmonella Typhimurium.
Proc.Natl.Acad.Sci.USA, 102, 2005
|
|
8XCN
 
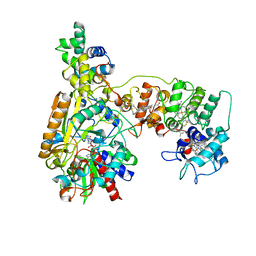 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1190A | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
8XCM
 
 | | Cryo-EM Structure of Membrane-bound Fructose Dehydrogenase from Gluconobacter japonicus variant-N1146Q | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Fructose dehydrogenase cytochrome subunit, ... | | Authors: | Fukawa, E, Miyata, T, Makino, F, Adachi, T, Suzuki, Y, Tanaka, H, Namba, K, Sowa, K, Kitazumi, Y, Shirai, O. | | Deposit date: | 2023-12-09 | | Release date: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural and electrochemical elucidation of biocatalytic mechanisms in direct electron transfer-type D-fructose dehydrogenase.
Electrochim Acta, 490, 2024
|
|
3MYU
 
 | | Mycoplasma genitalium MG289 | | Descriptor: | 3-(4-AMINO-2-METHYL-PYRIMIDIN-5-YLMETHYL)-5-(2-HYDROXY-ETHYL)-4-METHYL-THIAZOL-3-IUM, ACETATE ION, High affinity transport system protein p37 | | Authors: | Sippel, K.H, Boehlein, S.K, Govindasamy, L, Namiki, K, Satai, Y, Quirit, J.G, Agbandje-McKenna, M, Goodison, S, Rosser, C.J, Sankaran, B, McKenna, R. | | Deposit date: | 2010-05-11 | | Release date: | 2010-10-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into Mycoplasma genitalium metabolism revealed by the structure of MG289, an extracytoplasmic thiamine binding lipoprotein.
Proteins, 79, 2011
|
|
