7EQE
 
 | | Crystal Structure of a transcription factor | | Descriptor: | TetR/AcrR family transcriptional regulator | | Authors: | Uehara, S, Tsugita, A, Matsui, T, Yokoyama, T, Ostash, I, Ostash, B, Tanaka, Y. | | Deposit date: | 2021-05-01 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | The carbohydrate tail of landomycin A is responsible for its interaction with the repressor protein LanK.
Febs J., 289, 2022
|
|
6AL3
 
 | | Lys49 PLA2 BPII derived from the venom of Protobothrops flavoviridis. | | Descriptor: | Basic phospholipase A2 BP-II, SULFATE ION | | Authors: | Matsui, T, Kamata, S, Suzuki, A, Oda-Ueda, N, Ogawa, T, Tanaka, Y. | | Deposit date: | 2018-09-05 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | SDS-induced oligomerization of Lys49-phospholipase A2from snake venom.
Sci Rep, 9, 2019
|
|
3W9Z
 
 | | Crystal structure of DusC | | Descriptor: | FLAVIN MONONUCLEOTIDE, tRNA-dihydrouridine synthase C | | Authors: | Chen, M, Yu, J, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2013-04-19 | | Release date: | 2013-07-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of dihydrouridine synthase C (DusC) from Escherichia coli
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6ZBO
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in Complex with 1-(6-morpholinopyrimidin-4-yl)-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-5-ol (Molidustat) | | Descriptor: | 2-(6-morpholin-4-ylpyrimidin-4-yl)-4-(1,2,3-triazol-1-yl)pyrazol-3-ol, CHLORIDE ION, Egl nine homolog 1, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Holt-Martyn, J.P, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
6ZBN
 
 | | HIF Prolyl Hydroxylase 2 (PHD2/EGLN1) in complex with tert-butyl 6-(5-hydroxy-4-(1H-1,2,3-triazol-1-yl)-1H-pyrazol-1-yl)nicotinate (IOX4) | | Descriptor: | Egl nine homolog 1, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Figg Jr, W.D, McDonough, M.A, Nakashima, Y, Schofield, C.J. | | Deposit date: | 2020-06-08 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural Basis of Prolyl Hydroxylase Domain Inhibition by Molidustat.
Chemmedchem, 16, 2021
|
|
2ZKE
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), DNA (5'-D(*DGP*DCP*DAP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DAP*DG)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZKF
 
 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | DNA (5'-D(*DCP*DTP*DAP*DTP*DCP*(5CM)P*DGP*DGP*DTP*DGP*DA)-3'), DNA (5'-D(P*DCP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DAP*DGP*DA)-3'), E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
3VSE
 
 | | Crystal structure of methyltransferase | | Descriptor: | Putative uncharacterized protein, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Kita, S, Tanaka, Y, Yao, M, Tanaka, I. | | Deposit date: | 2012-04-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal structure of a putative methyltransferase SAV1081 from Staphylococcus aureus
Protein Pept.Lett., 20, 2012
|
|
3VZP
 
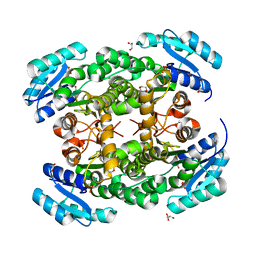 | | Crystal structure of PhaB from Ralstonia eutropha | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Acetoacetyl-CoA reductase, GLYCEROL, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.792 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
3VZS
 
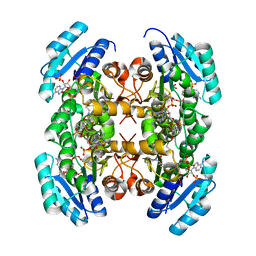 | | Crystal structure of PhaB from Ralstonia eutropha in complex with Acetoacetyl-CoA and NADP | | Descriptor: | ACETOACETYL-COENZYME A, Acetoacetyl-CoA reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Ikeda, K, Tanaka, Y, Tanaka, I, Yao, M. | | Deposit date: | 2012-10-15 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Directed evolution and structural analysis of NADPH-dependent Acetoacetyl Coenzyme A (Acetoacetyl-CoA) reductase from Ralstonia eutropha reveals two mutations responsible for enhanced kinetics
Appl.Environ.Microbiol., 79, 2013
|
|
2ZKD
 
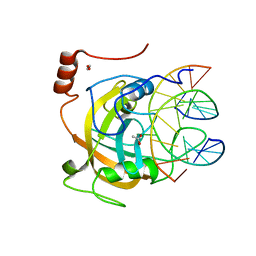 | | Crystal structure of the SRA domain of mouse Np95 in complex with hemi-methylated CpG DNA | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA (5'-D(*DCP*DTP*DAP*DCP*DCP*DGP*DGP*DAP*DTP*DTP*DGP*DC)-3'), ... | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
2ZKG
 
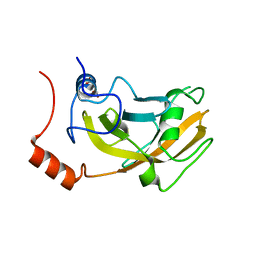 | | Crystal structure of unliganded SRA domain of mouse Np95 | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase UHRF1 | | Authors: | Arita, K, Ariyoshi, M, Tochio, H, Nakamura, Y, Shirakawa, M. | | Deposit date: | 2008-03-19 | | Release date: | 2008-09-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Recognition of hemi-methylated DNA by the SRA protein UHRF1 by a base-flipping mechanism
Nature, 455, 2008
|
|
4MMH
 
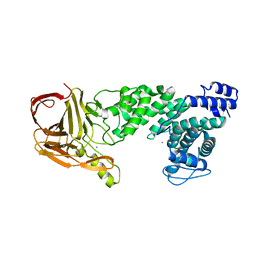 | | Crystal structure of heparan sulfate lyase HepC from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
4MMI
 
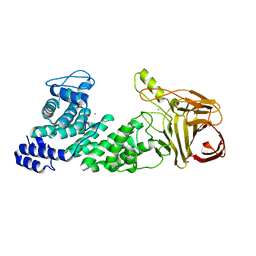 | | Crystal structure of heparan sulfate lyase HepC mutant from Pedobacter heparinus | | Descriptor: | CALCIUM ION, Heparinase III protein | | Authors: | Maruyama, Y, Nakamichi, Y, Mikami, B, Murata, K, Hashimoto, W. | | Deposit date: | 2013-09-09 | | Release date: | 2014-01-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pedobacter heparinus Heparin Lyase Hep III with the Active Site in a Deep Cleft
Biochemistry, 53, 2014
|
|
1WOZ
 
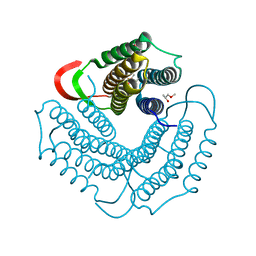 | | Crystal structure of uncharacterized protein ST1454 from Sulfolobus tokodaii | | Descriptor: | (20S)-2,5,8,11,14,17-HEXAMETHYL-3,6,9,12,15,18-HEXAOXAHENICOSANE-1,20-DIOL, 177aa long conserved hypothetical protein (ST1454) | | Authors: | Sasaki, T, Tanaka, Y, Yasutake, Y, Yao, M, Tanaka, I, Tsumoto, K, Kumagai, I. | | Deposit date: | 2004-08-27 | | Release date: | 2005-10-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Crystal structure of the uncharacterized protein ST1454 from Sulfolobus tokodaii.
To be Published
|
|
2ZKL
 
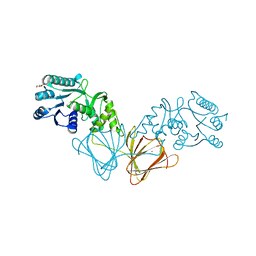 | | Crystal Structure of capsular polysaccharide assembling protein CapF from staphylococcus aureus | | Descriptor: | Capsular polysaccharide synthesis enzyme Cap5F, GLYCEROL, ZINC ION | | Authors: | Miyafusa, T, Tanaka, Y, Yao, M, Tanaka, I, Tsumoto, K. | | Deposit date: | 2008-03-25 | | Release date: | 2009-03-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Crystal structure of capsular polysaccharide assembling protein from Staphylococcus aureus
to be published
|
|
3AQA
 
 | | Crystal structure of the human BRD2 BD1 bromodomain in complex with a BRD2-interactive compound, BIC1 | | Descriptor: | 1-[2-(1H-benzimidazol-2-ylsulfanyl)ethyl]-3-methyl-1,3-dihydro-2H-benzimidazole-2-thione, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Bromodomain-containing protein 2 | | Authors: | Umehara, T, Nakamura, Y, Terada, T, Shirouzu, M, Padmanabhan, B, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-10-27 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Real-Time Imaging of Histone H4K12-Specific Acetylation Determines the Modes of Action of Histone Deacetylase and Bromodomain Inhibitors
Chem.Biol., 18, 2011
|
|
2YVZ
 
 | | Crystal structure of magnesium transporter MgtE cytosolic domain, Mg2+-free form | | Descriptor: | Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
2Z92
 
 | | Crystal structure of the Fab fragment of anti-ciguatoxin antibody 10C9 in complex with CTX3C_ABCDE | | Descriptor: | (4Z)-2,8:7,12:11,15:14,18:17,22-PENTAANHYDRO-4,5,6,9,10,13,19,20,21-NONADEOXY-D-ARABINO-D-ALLO-D-ALLO-DOCOSA-4,9,20-TRIENITOL, Anti-ciguatoxin antibody 10C9 Fab heavy chain, Anti-ciguatoxin antibody 10C9 Fab light chain, ... | | Authors: | Ui, M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2007-09-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How Protein Recognizes Ladder-like Polycyclic Ethers: INTERACTIONS BETWEEN CIGUATOXIN (CTX3C) FRAGMENTS AND ITS SPECIFIC ANTIBODY 10C9
J.Biol.Chem., 283, 2008
|
|
2Z91
 
 | | Crystal structure of the Fab fragment of anti-ciguatoxin antibody 10C9 | | Descriptor: | Anti-ciguatoxin antibody 10C9 FAB heavy chain, Anti-ciguatoxin antibody 10C9 FAB light chain | | Authors: | Ui, M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2007-09-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | How Protein Recognizes Ladder-like Polycyclic Ethers: INTERACTIONS BETWEEN CIGUATOXIN (CTX3C) FRAGMENTS AND ITS SPECIFIC ANTIBODY 10C9
J.Biol.Chem., 283, 2008
|
|
2Z93
 
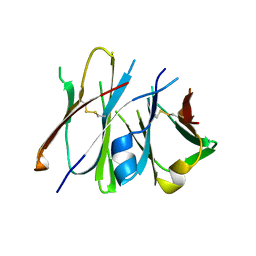 | | Crystal structure of Fab fragment of anti-ciguatoxin antibody 10C9 in complex with CTX3C-ABCD | | Descriptor: | 1,6:5,9:8,12:11,16-TETRAANHYDRO-2,3,4,10,13,14-HEXADEOXY-D-GLYCERO-D-ALLO-D-GULO-HEPTADECA-2,13-DIENITOL, Anti-ciguatoxin antibody 10C9 Fab heavy chain, Anti-ciguatoxin antibody 10C9 Fab light chain | | Authors: | Ui, M, Tanaka, Y, Tsumoto, K. | | Deposit date: | 2007-09-14 | | Release date: | 2008-05-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | How Protein Recognizes Ladder-like Polycyclic Ethers: INTERACTIONS BETWEEN CIGUATOXIN (CTX3C) FRAGMENTS AND ITS SPECIFIC ANTIBODY 10C9
J.Biol.Chem., 283, 2008
|
|
1UD9
 
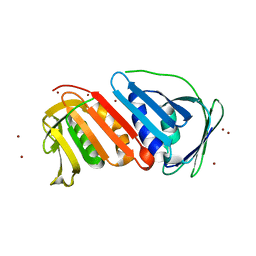 | | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii | | Descriptor: | DNA polymerase sliding clamp A, ZINC ION | | Authors: | Tanabe, E, Yasutake, Y, Tanaka, Y, Yao, M, Tsumoto, K, Kumagai, I, Tanaka, I. | | Deposit date: | 2003-04-28 | | Release date: | 2004-06-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Proliferating Cell Nuclear Antigen (PCNA) Homolog From Sulfolobus tokodaii
To be published
|
|
2DG7
 
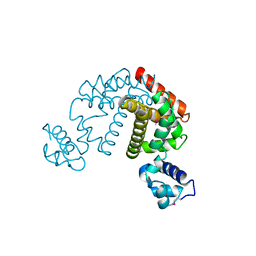 | | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2) | | Descriptor: | putative transcriptional regulator | | Authors: | Hayashi, T, Tanaka, Y, Sakai, N, Yao, M, Tamura, T, Tanaka, I. | | Deposit date: | 2006-03-08 | | Release date: | 2007-03-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the putative transcriptional regulator SCO0337 from Streptomyces coelicolor A3(2)
To be Published
|
|
2RVP
 
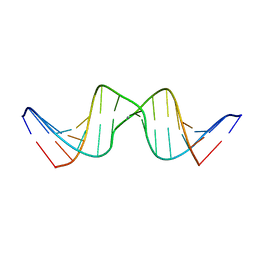 | | Solution structure of DNA Containing Metallo-Base-Pair | | Descriptor: | DNA (5'-D(*TP*AP*AP*TP*AP*TP*AP*CP*TP*TP*AP*AP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*TP*TP*AP*AP*CP*TP*AP*TP*AP*TP*TP*A)-3'), SILVER ION | | Authors: | Dairaku, T, Furuita, K, Sato, H, Sebera, J, Nakashima, K, Kondo, J, Yamanaka, D, Kondo, Y, Okamoto, I, Ono, A, Sychrovsky, V, Kojima, C, Tanaka, Y. | | Deposit date: | 2016-03-22 | | Release date: | 2016-08-31 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure Determination of an Ag(I) -Mediated Cytosine-Cytosine Base Pair within DNA Duplex in Solution with (1) H/(15) N/(109) Ag NMR Spectroscopy.
Chemistry, 22, 2016
|
|
3A0A
 
 | | Structure of (PPG)4-OPG-(PPG)4, monoclinic, twinned crystal | | Descriptor: | collagen-like peptide | | Authors: | Okuyama, K, Morimoto, T, Hongo, C, Katagiri, A, Tanaka, Y, Nishino, N. | | Deposit date: | 2009-03-13 | | Release date: | 2010-03-09 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Crystal structure of (PPG)4-OPG-(PPG)4
To be Published
|
|
