6FQE
 
 | | Phosphotriesterase PTE_A53_4 | | Descriptor: | (4~{S},6~{R})-2,2,6-trimethyl-1,3-dioxan-4-ol, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Phosphotriesterase
PTE_A53_4
To Be Published
|
|
6FMX
 
 | | IMISX-EP of W-PgpB | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Phosphatidylglycerophosphatase B, TUNGSTATE(VI)ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6P7L
 
 | |
6CHH
 
 | | Structure of human NNMT in complex with bisubstrate inhibitor MS2756 | | Descriptor: | (2~{S})-5-[2-(3-aminocarbonylphenyl)ethyl-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]amino]-2-azanyl-pentanoic acid, 1,2-ETHANEDIOL, Nicotinamide N-methyltransferase | | Authors: | Babault, N, Liu, J, Jin, J. | | Deposit date: | 2018-02-22 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovery of Bisubstrate Inhibitors of Nicotinamide N-Methyltransferase (NNMT).
J. Med. Chem., 61, 2018
|
|
6V06
 
 | | Crystal structure of Beta-2 glycoprotein I purified from plasma (pB2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-beta-D-mannopyranose-(1-6)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-2-glycoprotein 1, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
5D4I
 
 | | Intact nitrite complex of a copper nitrite reductase determined by serial femtosecond crystallography | | Descriptor: | COPPER (II) ION, Copper-containing nitrite reductase, NITRITE ION | | Authors: | Fukuda, Y, Tse, K.M, Nakane, T, Nakatsu, T, Suzuki, M, Sugahara, M, Inoue, S, Masuda, T, Yumoto, F, Matsugaki, N, Nango, E, Tono, K, Joti, Y, Kameshima, T, Song, C, Hatsui, T, Yabashi, M, Nureki, O, Murphy, M.E.P, Inoue, T, Iwata, S, Mizohata, E. | | Deposit date: | 2015-08-07 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Redox-coupled proton transfer mechanism in nitrite reductase revealed by femtosecond crystallography
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6V09
 
 | | Crystal structure of human recombinant Beta-2 glycoprotein I short tag (ST-B2GPI) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-glycoprotein 1, SULFATE ION, ... | | Authors: | Chen, Z, Ruben, E.A, Planer, W, Chinnaraj, M, Zuo, X, Pengo, V, Macor, P, Tedesco, F, Pozzi, N. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | The J-elongated conformation of beta2-glycoprotein I predominates in solution: implications for our understanding of antiphospholipid syndrome.
J.Biol.Chem., 295, 2020
|
|
6V0F
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 4 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6FQF
 
 | | THE X-RAY STRUCTURE OF FERRIC-BETA4 HUMAN HEMOGLOBIN | | Descriptor: | Hemoglobin subunit beta, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Mazzarella, L, Merlino, A, Balasco, N, Balsamo, A, Vergara, A. | | Deposit date: | 2018-02-14 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the ferric homotetrameric beta4human hemoglobin.
Biophys. Chem., 240, 2018
|
|
1R7E
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure. Sample in 100mM SDS). | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
6V0H
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 6 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
5CUB
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 314268-40-1 (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, methyl 4-[(4-methylpiperazin-1-yl)methyl]benzoate | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 314268-40-1 (SGC - Diamond I04-1 fragment screening)
To be published
|
|
5CUG
 
 | | Crystal structure of the bromodomain of bromodomain adjacent to zinc finger domain protein 2B (BAZ2B) in complex with 4-Acetamidobenzoic acid (SGC - Diamond I04-1 fragment screening) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B, PARA ACETAMIDO BENZOIC ACID | | Authors: | Bradley, A, Pearce, N, Krojer, T, Ng, J, Talon, R, Vollmar, M, Jose, B, von Delft, F, Bountra, C, Arrowsmith, C.H, Edwards, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-07-24 | | Release date: | 2015-09-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of the second bromodomain of bromodomain adjancent to zinc finger domain protein 2B (BAZ2B) in complex with 4-Acetamidobenzoic acid (SGC - Diamond I04-1 fragment screening)
To be published
|
|
6V11
 
 | | Lon Protease from Yersinia pestis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Lon protease | | Authors: | Shin, M, Puchades, C, Asmita, A, Puri, N, Adjei, E, Wiseman, R.L, Karzai, A.W, Lander, G.C. | | Deposit date: | 2019-11-19 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for distinct operational modes and protease activation in AAA+ protease Lon.
Sci Adv, 6, 2020
|
|
1R1W
 
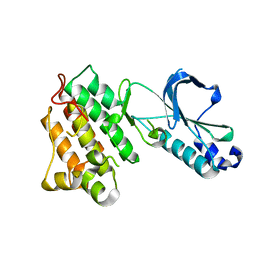 | | CRYSTAL STRUCTURE OF THE TYROSINE KINASE DOMAIN OF THE HEPATOCYTE GROWTH FACTOR RECEPTOR C-MET | | Descriptor: | HEPATOCYTE GROWTH FACTOR RECEPTOR | | Authors: | Schiering, N, Knapp, S, Marconi, M, Flocco, M.M, Cui, J, Perego, R, Rusconi, L, Cristiani, C. | | Deposit date: | 2003-09-25 | | Release date: | 2003-10-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the tyrosine kinase domain of the hepatocyte growth factor receptor c-Met and its complex with the microbial alkaloid K-252a
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
5CX6
 
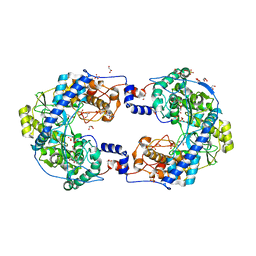 | | Crystal structure of Thosea asigna virus RNA-dependent RNA polymerase (RdRP) complexed with CDP | | Descriptor: | CYTIDINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-07-28 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
1R2C
 
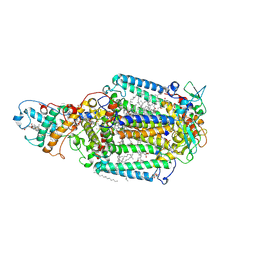 | | PHOTOSYNTHETIC REACTION CENTER BLASTOCHLORIS VIRIDIS (ATCC) | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Baxter, R.H, Ponomarenko, N, Pahl, R, Srajer, V, Moffat, K, Norris, J.R. | | Deposit date: | 2003-09-26 | | Release date: | 2004-04-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.86 Å) | | Cite: | Time-resolved crystallographic studies of light-induced structural changes in the photosynthetic reaction center.
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
6FS3
 
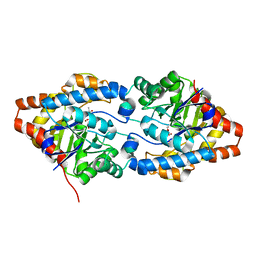 | | Phosphotriesterase PTE_A53_1 | | Descriptor: | (2~{S})-2-methylpentanedioic acid, FORMIC ACID, Parathion hydrolase, ... | | Authors: | Dym, O, Aggarwal, N, Albeck, S, Unger, T, Hamer Rogotner, S, Silman, I, Leader, H, Ashani, Y, Goldsmith, M, Greisen, P, Tawfik, D, Sussman, L.J. | | Deposit date: | 2018-02-19 | | Release date: | 2019-03-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structures of Bacterail Phosphotriesterase variant with high catalytic activity towards organophosphate nerve agents developed by use of structure-based design and molecular evolution
To Be Published
|
|
1A54
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C LABELLED WITH A COUMARIN FLUOROPHORE AND BOUND TO DIHYDROGENPHOSPHATE ION | | Descriptor: | DIHYDROGENPHOSPHATE ION, N-[2-(1-MALEIMIDYL)ETHYL]-7-DIETHYLAMINOCOUMARIN-3-CARBOXAMIDE, Phosphate-binding protein PstS | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
6PK6
 
 | | Human PRPF4B bound to benzothiophene inhibitor 329 | | Descriptor: | 4-(5-{[(2-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, SULFATE ION, Serine/threonine-protein kinase PRP4 homolog | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | to be published
To Be Published
|
|
5CYR
 
 | | Crystal structure of Thosea asigna virus RNA-dependent RNA polymerase (RdRP) complexed with ATP and ssRNA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-07-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.504 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
1A55
 
 | | PHOSPHATE-BINDING PROTEIN MUTANT A197C | | Descriptor: | DIHYDROGENPHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Hirshberg, M, Henrick, K, Lloyd-Haire, L, Vasisht, N, Brune, M, Corrie, J.E.T, Webb, M.R. | | Deposit date: | 1998-02-19 | | Release date: | 1998-10-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphate binding protein labeled with a coumarin fluorophore, a probe for inorganic phosphate.
Biochemistry, 37, 1998
|
|
6FTZ
 
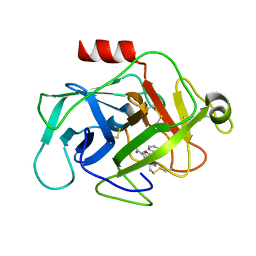 | | COMPLEMENT FACTOR D COMPLEXED WITH COMPOUND 6 | | Descriptor: | Complement factor D, ~{N}4-[3-(aminomethyl)phenyl]-1~{H}-indole-2,4-dicarboxamide | | Authors: | Ostermann, N. | | Deposit date: | 2018-02-26 | | Release date: | 2018-06-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
1R7S
 
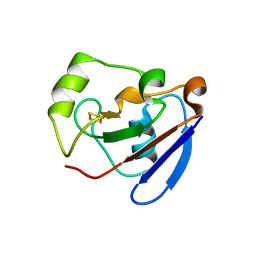 | | PUTIDAREDOXIN (Fe2S2 ferredoxin), C73G mutant | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, Putidaredoxin | | Authors: | Smith, N, Mayhew, M, Kelly, H, Robinson, H, Heroux, A, Holden, M.J, Gallagher, D.T. | | Deposit date: | 2003-10-22 | | Release date: | 2004-04-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Structure of C73G putidaredoxin from Pseudomonas putida.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
6D2X
 
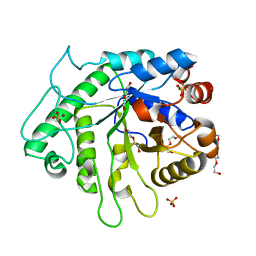 | | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36,39-TRIDECAOXAHENTETRACONTANE-1,41-DIOL, Aryl-phospho-beta-D-glucosidase BglC, GH1 family, ... | | Authors: | Stogios, P.J, Skarina, T, McGregor, N, Di Leo, R, Brumer, H, Savchenko, A. | | Deposit date: | 2018-04-14 | | Release date: | 2019-10-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Crystal structure of the GH26 domain from PbGH26-GH5A endo-beta-mannanase/endo-beta-glucanase from Prevotella bryantii
To Be Published
|
|
