1WIY
 
 | | Crystal Structure Analysis of a 6-coordinated Cytochorome P450 from Thermus thermophilus HB8 | | Descriptor: | Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kousumi, Y, Nakagawa, N, Kaneko, M, Yamamoto, H, Masui, R, Kuramitsu, S, Ueyama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure Analysis of Cytochrome P450 from Thermus thermophilus HB8
To be Published
|
|
4BG6
 
 | | 14-3-3 interaction with Rnd3 prenyl-phosphorylation motif | | Descriptor: | 14-3-3 PROTEIN ZETA/DELTA, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, FARNESYL, ... | | Authors: | Riou, P, Kjaer, S, Purkiss, A, O'Reilly, N, McDonald, N.Q. | | Deposit date: | 2013-03-23 | | Release date: | 2013-05-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 14-3-3 Proteins Interact with a Hybrid Prenyl-Phosphorylation Motif to Inhibit G Proteins.
Cell(Cambridge,Mass.), 153, 2013
|
|
1WU7
 
 | | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum | | Descriptor: | Histidyl-tRNA synthetase | | Authors: | Tanaka, Y, Sakai, N, Yao, M, Watanabe, N, Tamura, T, Tanaka, I. | | Deposit date: | 2004-12-01 | | Release date: | 2005-12-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of histidyl-tRNA synthetase from Thermoplasma acidophilum
To be Published
|
|
5TCG
 
 | | Crystal structure of tryptophan synthase from M. tuberculosis - aminoacrylate-bound form | | Descriptor: | 2-[({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)AMINO]ACRYLIC ACID, CESIUM ION, FORMIC ACID, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
1WRB
 
 | | Crystal structure of the N-terminal RecA-like domain of DjVLGB, a pranarian Vasa-like RNA helicase | | Descriptor: | DjVLGB, SULFATE ION | | Authors: | Kurimoto, K, Muto, Y, Obayashi, N, Terada, T, Shirouzu, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Kigawa, T, Okumura, H, Tanaka, A, Shibata, N, Kashikawa, M, Agata, K, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-10-14 | | Release date: | 2005-04-12 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the N-terminal RecA-like domain of a DEAD-box RNA helicase, the Dugesia japonica vasa-like gene B protein
J.Struct.Biol., 150, 2005
|
|
5XBU
 
 | | Crystal structure of GH45 endoglucanase EG27II in apo-form | | Descriptor: | Endo-beta-1,4-glucanase | | Authors: | Nomura, T, Mizutani, K, Iwase, H, Takahashi, N, Mikami, B. | | Deposit date: | 2017-03-21 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | High-resolution crystal structures of the glycoside hydrolase family 45 endoglucanase EG27II from the snail Ampullaria crossean.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
1WGP
 
 | | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel | | Descriptor: | Probable cyclic nucleotide-gated ion channel 6 | | Authors: | Chikayama, E, Nameki, N, Kigawa, T, Koshiba, S, Inoue, M, Tomizawa, T, Kobayashi, N, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-28 | | Release date: | 2004-11-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the cNMP-binding domain from Arabidopsis thaliana cyclic nucleotide-regulated ion channel
To be Published
|
|
4BHY
 
 | | Structure of alanine racemase from Aeromonas hydrophila | | Descriptor: | ALANINE RACEMASE | | Authors: | Otero, L.H, Carrasco-Lopez, C, Bernardo-Garcia, N, Hermoso, J.A. | | Deposit date: | 2013-04-09 | | Release date: | 2014-01-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural Basis for the Broad Specificity of a New Family of Amino-Acid Racemases.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
1WWU
 
 | | Solution structure of the SAM_PNT domain of human protein FLJ21935 | | Descriptor: | Hypothetical protein FLJ21935 | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT domain of human protein FLJ21935
To be Published
|
|
1EU2
 
 | |
5TCF
 
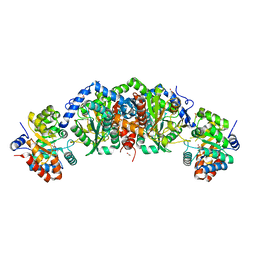 | | Crystal structure of tryptophan synthase from M. tuberculosis - ligand-free form | | Descriptor: | FORMIC ACID, MALONATE ION, POTASSIUM ION, ... | | Authors: | Michalska, K, Maltseva, N, Jedrzejczak, R, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-15 | | Release date: | 2017-05-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | A small-molecule allosteric inhibitor of Mycobacterium tuberculosis tryptophan synthase.
Nat. Chem. Biol., 13, 2017
|
|
1EU6
 
 | |
5YE1
 
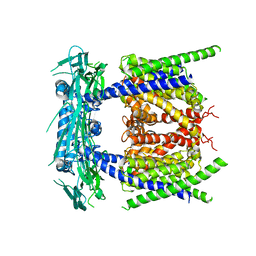 | |
5YHP
 
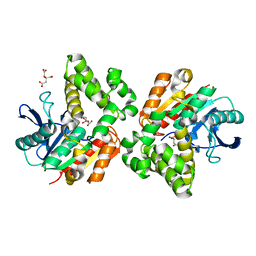 | | Proline iminopeptidase from Psychrophilic yeast glaciozyma antarctica | | Descriptor: | CITRATE ANION, Cold active proline iminopeptidase | | Authors: | Rodzli, N.A, Kamaruddin, S, Jonet, A, Seman, W.M.K.W, Tab, M.M, Minor, N, Jaafar, N.R, Mahadi, N.M, Murad, A.M.A, Bakar, F.D.A, Illias, R.M.D. | | Deposit date: | 2017-09-29 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Biochemical Characterisation and Structure Determination of a Novel Cold-Active Proline Iminopeptidase from the Psychrophilic Yeast, Glaciozyma antarctica PI12
Catalysts, 12, 2022
|
|
7MCD
 
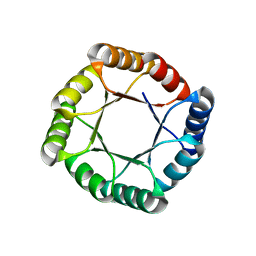 | |
7MCC
 
 | | Crystal structure of an AI-designed TIM-barrel F2C | | Descriptor: | AI-designed TIM-barrel F2C, SULFATE ION | | Authors: | Mathews, I.I, Anand-Achim, N, Perez, C.P, Huang, P.S. | | Deposit date: | 2021-04-02 | | Release date: | 2022-01-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Protein sequence design with a learned potential.
Nat Commun, 13, 2022
|
|
1EMR
 
 | | CRYSTAL STRUCTURE OF HUMAN LEUKEMIA INHIBITORY FACTOR (LIF) | | Descriptor: | LEUKEMIA INHIBITORY FACTOR | | Authors: | Robinson, R.C, Heath, J.K, Hawkins, N, Samal, B, Jones, E.Y, Betzel, C. | | Deposit date: | 2000-03-17 | | Release date: | 2001-03-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Species Variation in Receptor Binding Site Revealed by the Medium Resolution X-ray Structure of Human Leukemia Inhibitory Factor
to be published
|
|
5Y48
 
 | | Crystal structure of the complex of Ribosome inactivating protein from Momordica balsamina with Pyrimidine-2,4-dione at 1.70 Angstrom resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Ribosome inactivating protein, URACIL | | Authors: | Singh, P.K, Pandey, S, Iqbal, N, Kaur, P, Sharma, S, Singh, T.P. | | Deposit date: | 2017-08-01 | | Release date: | 2017-09-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Binding and structural studies of the complexes of type 1 ribosome inactivating protein from Momordica balsamina with uracil and uridine.
Proteins, 87, 2019
|
|
6J4P
 
 | |
6J4V
 
 | | Structural basis of tubulin detyrosination by vasohibins-SVBP enzyme complex and functional implications | | Descriptor: | GLYCEROL, PHOSPHATE ION, Small vasohibin-binding protein, ... | | Authors: | Wang, N, Bao, H, Huang, H. | | Deposit date: | 2019-01-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of tubulin detyrosination by the vasohibin-SVBP enzyme complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
4CAD
 
 | | Mechanism of farnesylated CAAX protein processing by the integral membrane protease Rce1 | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Kulkarni, K, Manolaridis, I, Dodd, R.B, Cronin, N, Ogasawara, S, Iwata, S, Barford, D. | | Deposit date: | 2013-10-08 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Farnesylated Caax Protein Processing by the Intramembrane Protease Rce1
Nature, 504, 2013
|
|
1ESW
 
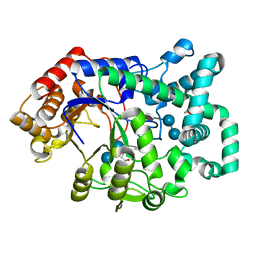 | | X-RAY STRUCTURE OF ACARBOSE BOUND TO AMYLOMALTASE FROM THERMUS AQUATICUS. IMPLICATIONS FOR THE SYNTHESIS OF LARGE CYCLIC GLUCANS | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, AMYLOMALTASE | | Authors: | Przylas, I, Terada, Y, Fujii, K, Takaha, T, Saenger, W, Straeter, N. | | Deposit date: | 2000-04-11 | | Release date: | 2001-04-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | X-ray structure of acarbose bound to amylomaltase from Thermus aquaticus. Implications for the synthesis of large cyclic glucans.
Eur.J.Biochem., 267, 2000
|
|
1WK1
 
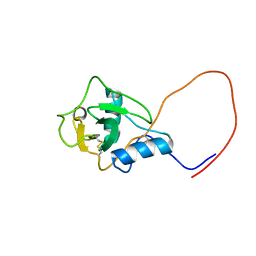 | | Solution structure of Lectin C-type domain derived from a hypothetical protein from C. elegans | | Descriptor: | Hypothetical protein yk1067a12 | | Authors: | Kobayashi, N, Koshiba, S, Inoue, M, Tochio, N, Kigawa, T, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-29 | | Release date: | 2004-11-29 | | Last modified: | 2022-03-02 | | Method: | SOLUTION NMR | | Cite: | Solution structure of Lectin C-type domain derived from a hypothetical protein from C. elegans
To be Published
|
|
5Y25
 
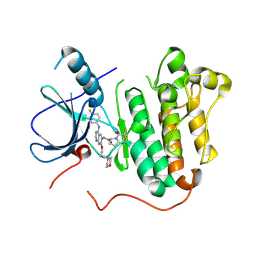 | | EGFR kinase domain mutant (T790M/L858R) with covalent ligand NS-062 | | Descriptor: | (2R)-N-[4-[(3-chloranyl-4-fluoranyl-phenyl)amino]-7-(3-morpholin-4-ylpropoxy)quinazolin-6-yl]-1-(2-fluoranylethanoyl)pyrrolidine-2-carboxamide, Epidermal growth factor receptor | | Authors: | Shiroishi, M, Abe, Y, Caaveiro, J.M.M, Sakamoto, S, Morimoto, S, Fuchida, H, Shindo, N, Ojida, A. | | Deposit date: | 2017-07-24 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.102 Å) | | Cite: | Selective and reversible modification of kinase cysteines with chlorofluoroacetamides.
Nat.Chem.Biol., 15, 2019
|
|
7DNS
 
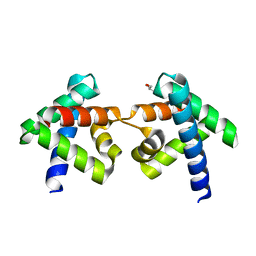 | | Crystal structure of domain-swapped dimer of H5_Fold-0 Elsa; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | GLYCEROL, de novo designed protein | | Authors: | Suzuki, K, Kobayashi, N, Murata, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-12-10 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.327 Å) | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
