4ROC
 
 | | Human TFIIB-related factor 2 (Brf2) and TBP bound to U6#2 promoter | | Descriptor: | MAGNESIUM ION, Non-template strand, TATA-box-binding protein, ... | | Authors: | Vannini, A, Gouge, J, Satia, K, Guthertz, N. | | Deposit date: | 2014-10-28 | | Release date: | 2015-12-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redox Signaling by the RNA Polymerase III TFIIB-Related Factor Brf2.
Cell(Cambridge,Mass.), 163, 2015
|
|
4RUP
 
 | | Crystal structure of zVDR L337H mutant-Gemini72 complex | | Descriptor: | (1R,3R,7E,17beta)-17-[(1R)-6,6,6-trifluoro-5-hydroxy-1-(4-hydroxy-4-methylpentyl)-5-(trifluoromethyl)hex-3-yn-1-yl]-9,1 0-secoestra-5,7-diene-1,3-diol, Nuclear receptor coactivator 1, Vitamin D3 receptor A | | Authors: | Huet, T, Moras, D, Rochel, N. | | Deposit date: | 2014-11-21 | | Release date: | 2015-10-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A vitamin D receptor selectively activated by gemini analogs reveals ligand dependent and independent effects.
Cell Rep, 10, 2015
|
|
6N9T
 
 | | Structure of a peptide-based photo-affinity cross-linker with Herceptin Fc | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin G1 FC, ... | | Authors: | Sadowsky, J, Ultsch, M, Vance, N, Wang, W. | | Deposit date: | 2018-12-04 | | Release date: | 2019-01-16 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.576 Å) | | Cite: | Development, Optimization, and Structural Characterization of an Efficient Peptide-Based Photoaffinity Cross-Linking Reaction for Generation of Homogeneous Conjugates from Wild-Type Antibodies.
Bioconjug. Chem., 30, 2019
|
|
4WZM
 
 | | Mutant K18E of RNA dependent RNA polymerase from Foot-and-Mouth Disease Virus complexed with RNA | | Descriptor: | ACETATE ION, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Verdaguer, N, Ferrer-Orta, C, de la Higuera, N. | | Deposit date: | 2014-11-20 | | Release date: | 2015-05-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Multifunctionality of a picornavirus polymerase domain: nuclear localization signal and nucleotide recognition.
J.Virol., 89, 2015
|
|
4TWZ
 
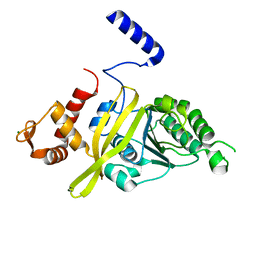 | | Crystal Structure Analysis of E Coli. RecA Protein | | Descriptor: | MAGNESIUM ION, Protein RecA | | Authors: | Hikima, T, Hiraki, T, Furuse, M, Ikawa, S, Iwasaki, W, Shibata, T, Kamiya, N. | | Deposit date: | 2014-07-02 | | Release date: | 2015-07-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Loop L1 governs the DNA-binding specificity and order for RecA-catalyzed reactions in homologous recombination and DNA repair
Nucleic Acids Res., 43, 2015
|
|
6YJM
 
 | | Crystal Structure of the Catalytic Domain of ADAMTS-5 in Complex with the Inhibitor GLPG1972 | | Descriptor: | (5~{S})-5-[3-[(3~{S})-4-[3,5-bis(fluoranyl)phenyl]-3-methyl-piperazin-1-yl]-3-oxidanylidene-propyl]-5-cyclopropyl-imidazolidine-2,4-dione, A disintegrin and metalloproteinase with thrombospondin motifs 5, CALCIUM ION, ... | | Authors: | Goepfert, A, Leonard, P, Triballeau, N, Fleury, D, Mollat, P, Lamers, M. | | Deposit date: | 2020-04-03 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Discovery of GLPG1972/S201086, a Potent, Selective, and Orally Bioavailable ADAMTS-5 Inhibitor for the Treatment of Osteoarthritis.
J.Med.Chem., 64, 2021
|
|
7T62
 
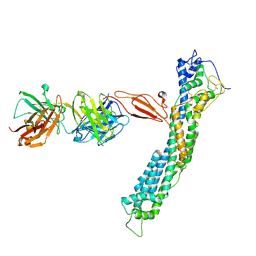 | | GPC2 HEP CT3 complex | | Descriptor: | CT3, Glypican-2 | | Authors: | Zhu, J, Cachau, R, De Val Alda, N, Li, N, Ho, M. | | Deposit date: | 2021-12-13 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (21 Å) | | Cite: | CAR T cells targeting tumor-associated exons of glypican 2 regress neuroblastoma in mice.
Cell Rep Med, 2, 2021
|
|
1BXE
 
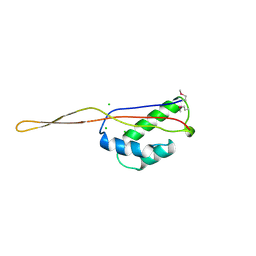 | | RIBOSOMAL PROTEIN L22 FROM THERMUS THERMOPHILUS | | Descriptor: | CHLORIDE ION, PROTEIN (RIBOSOMAL PROTEIN L22) | | Authors: | Unge, J, Aberg, A, Al-Karadaghi, S, Nikulin, A, Nikonov, S, Davydova, N, Nevskaya, N, Garber, M, Liljas, A. | | Deposit date: | 1998-10-02 | | Release date: | 1998-10-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The crystal structure of ribosomal protein L22 from Thermus thermophilus: insights into the mechanism of erythromycin resistance.
Structure, 6, 1998
|
|
7WMT
 
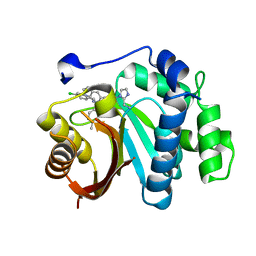 | | Crystal structure of small molecule 13 bound to human Nicotinamide N-methyltransferase | | Descriptor: | Nicotinamide N-methyltransferase, [(2~{R},4~{S})-4-[2-(aminomethyl)imidazol-1-yl]-2-[1-[(4-chlorophenyl)methyl]-5-methyl-indol-2-yl]pyrrolidin-1-yl]-(1~{H}-pyrrolo[2,3-b]pyridin-5-yl)methanone | | Authors: | Yoshida, S, Uehara, S, Kondo, N, Takahashi, Y, Yamamoto, S, Kameda, A, Kawagoe, S, Inoue, N, Yamada, M, Yoshimura, N, Tachibana, Y. | | Deposit date: | 2022-01-17 | | Release date: | 2022-08-31 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Peptide-to-Small Molecule: A Pharmacophore-Guided Small Molecule Lead Generation Strategy from High-Affinity Macrocyclic Peptides.
J.Med.Chem., 65, 2022
|
|
5LME
 
 | | Specific-DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase | | Descriptor: | ZINC ION, piggyBac transposase | | Authors: | Morellet, N, Taylor, J.A, Wieninger, S, Moriau, S, Li, X, Lescop, E, Mathy, N, Bischerour, J, Betermier, M, Bardiaux, B, Nilges, M, Craig, N.L, Hickman, A.B, Dyda, F, Guittet, E. | | Deposit date: | 2016-07-30 | | Release date: | 2017-12-20 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Sequence-specific DNA binding activity of the cross-brace zinc finger motif of the piggyBac transposase.
Nucleic Acids Res., 46, 2018
|
|
7BQS
 
 | | Solution NMR structure of fold-U Nomur; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Nomur | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
7BQR
 
 | | Solution NMR structure of fold-K Mussoc; de novo designed protein with an asymmetric all-alpha topology | | Descriptor: | Mussoc | | Authors: | Kobayashi, N, Nagashima, T, Sakuma, K, Kosugi, T, Koga, R, Koga, N. | | Deposit date: | 2020-03-25 | | Release date: | 2021-04-07 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Design of complicated all-alpha protein structures
Nat.Struct.Mol.Biol., 2024
|
|
6QFI
 
 | | Structure of human Mcl-1 in complex with BIM BH3 peptide | | Descriptor: | Bcl-2-like protein 11, Induced myeloid leukemia cell differentiation protein Mcl-1, ZINC ION | | Authors: | Dokurno, P, Murray, J, Davidson, J, Chen, I, Davis, B, Graham, C.J, Harris, R, Jordan, A.M, Matassova, N, Pedder, C, Ray, S, Roughley, S, Smith, J, Walmsley, C, Wang, Y, Whitehead, N, Williamson, D.S, Casara, P, Le Diguarher, T, Hickman, J, Stark, J, Kotschy, A, Geneste, O, Hubbard, R.E. | | Deposit date: | 2019-01-10 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Establishing Drug Discovery and Identification of Hit Series for the Anti-apoptotic Proteins, Bcl-2 and Mcl-1.
Acs Omega, 4, 2019
|
|
6BRT
 
 | | F-box protein CTH with hydrolase | | Descriptor: | (5R)-5-hydroxy-3-methylfuran-2(5H)-one, D3-CTH-D14-D-ring | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-12-01 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
1GC2
 
 | | CRYSTAL STRUCTURE OF THE PYRIDOXAL-5'-PHOSPHATE DEPENDENT L-METHIONINE GAMMA-LYASE FROM PSEUDOMONAS PUTIDA | | Descriptor: | METHIONINE GAMMA-LYASE | | Authors: | Motoshima, H, Inagaki, K, Kumasaka, T, Furuichi, M, Inoue, H, Tamura, T, Esaki, N, Soda, K, Tanaka, N, Yamamoto, M, Tanaka, H. | | Deposit date: | 2000-07-06 | | Release date: | 2002-05-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the pyridoxal 5'-phosphate dependent L-methionine gamma-lyase from Pseudomonas putida.
J.Biochem., 128, 2000
|
|
6BRQ
 
 | | Crystal structure of rice ASK1-D3 ubiquitin ligase complex crystal form 3 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
5OF1
 
 | | The structural versatility of TasA in B. subtilis biofilm formation | | Descriptor: | 1,2-ETHANEDIOL, 2-HYDROXYBENZOIC ACID, Spore coat-associated protein N | | Authors: | Roske, Y, Diehl, A, Ball, L, Chowdhury, A, Hiller, M, Moliere, N, Kramer, R, Nagaraj, M, Stoeppler, D, Worth, C.L, Schlegel, B, Leidert, M, Cremer, N, Eisenmenger, F, Lopez, D, Schmieder, P, Heinemann, U, Turgay, K, Akbey, U, Oschkinat, H. | | Deposit date: | 2017-07-10 | | Release date: | 2018-03-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural changes of TasA in biofilm formation ofBacillus subtilis.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6NZ8
 
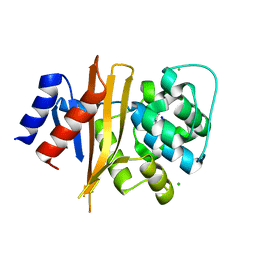 | | Structure of carbamylated apo OXA-231 carbapenemase | | Descriptor: | Beta-lactamase OXA-231, CHLORIDE ION, SODIUM ION | | Authors: | Favaro, D.C, Llontop, E.E, Vasconcelos, F.N, Antunes, V.U, Farah, S.C, Lincopan, N. | | Deposit date: | 2019-02-13 | | Release date: | 2020-02-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Importance of the beta 5-beta 6 Loop for the Structure, Catalytic Efficiency, and Stability of Carbapenem-Hydrolyzing Class D beta-Lactamase Subfamily OXA-143.
Biochemistry, 58, 2019
|
|
6DSZ
 
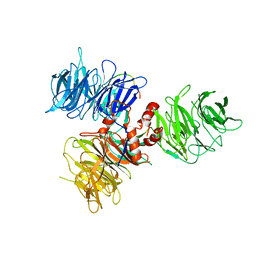 | |
1UMD
 
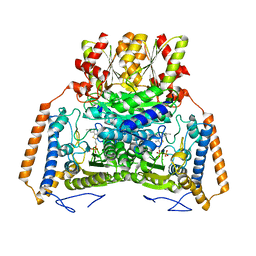 | | branched-chain 2-oxo acid dehydrogenase (E1) from Thermus thermophilus HB8 with 4-methyl-2-oxopentanoate as an intermediate | | Descriptor: | 2-OXO-4-METHYLPENTANOIC ACID, 2-oxo acid dehydrogenase alpha subunit, 2-oxo acid dehydrogenase beta subunit, ... | | Authors: | Nakai, T, Nakagawa, N, Maoka, N, Masui, R, Kuramitsu, S, Kamiya, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-09-25 | | Release date: | 2004-03-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Ligand-induced Conformational Changes and a Reaction Intermediate in Branched-chain 2-Oxo Acid Dehydrogenase (E1) from Thermus thermophilus HB8, as Revealed by X-ray Crystallography
J.Mol.Biol., 337, 2004
|
|
6BRP
 
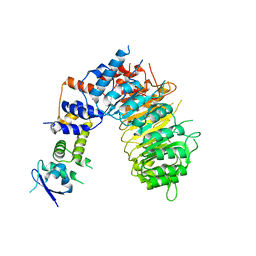 | | F-box protein form 2 | | Descriptor: | F-box/LRR-repeat MAX2 homolog, SKP1-like protein 1A | | Authors: | Shabek, N, Zheng, N, Mao, H, Hinds, T.R, Ticchiarelli, F, Leyser, O. | | Deposit date: | 2017-11-30 | | Release date: | 2018-11-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Structural plasticity of D3-D14 ubiquitin ligase in strigolactone signalling.
Nature, 563, 2018
|
|
1DIX
 
 | | CRYSTAL STRUCTURE OF RNASE LE | | Descriptor: | EXTRACELLULAR RIBONUCLEASE LE | | Authors: | Tanaka, N, Nakamura, K.T. | | Deposit date: | 1999-11-30 | | Release date: | 2000-09-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a plant ribonuclease, RNase LE.
J.Mol.Biol., 298, 2000
|
|
6BRO
 
 | |
1C51
 
 | | PHOTOSYNTHETIC REACTION CENTER AND CORE ANTENNA SYSTEM (TRIMERIC), ALPHA CARBON ONLY | | Descriptor: | CHLOROPHYLL A, IRON/SULFUR CLUSTER, PHYLLOQUINONE, ... | | Authors: | Klukas, O, Schubert, W.D, Jordan, P, Krauss, N, Fromme, P, Witt, H.T, Saenger, W. | | Deposit date: | 1999-10-21 | | Release date: | 2000-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Photosystem I, an improved model of the stromal subunits PsaC, PsaD, and PsaE.
J.Biol.Chem., 274, 1999
|
|
5I8L
 
 | | Crystal structure of the full-length cell wall-binding module of Cpl7 mutant R223A | | Descriptor: | GLYCEROL, Lysozyme | | Authors: | Bernardo-Garcia, N, Silva-Martin, N, Uson, I, Hermoso, J.A. | | Deposit date: | 2016-02-19 | | Release date: | 2017-03-08 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Deciphering how Cpl-7 cell wall-binding repeats recognize the bacterial peptidoglycan.
Sci Rep, 7, 2017
|
|
