6MVE
 
 | | Reduced X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-25 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6N91
 
 | | Crystal Structure of Adenosine Deaminase from Vibrio cholerae Complexed with Pentostatin (Deoxycoformycin) | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYCOFORMYCIN, 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, ... | | Authors: | Maltseva, N, Kim, Y, Endres, M, Welk, L, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-11-30 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Adenosine Deaminase from Vibrio cholerae Complexed with Pentostatin (Deoxycoformycin) (CASP target)
To Be Published
|
|
6N8V
 
 | | Hsp104DWB open conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
2CMM
 
 | | STRUCTURAL ANALYSIS OF THE MYOGLOBIN RECONSTITUTED WITH IRON PORPHINE | | Descriptor: | CYANIDE ION, MYOGLOBIN, PORPHYRIN FE(III) | | Authors: | Sato, T, Tanaka, N, Moriyama, H, Igarashi, N, Neya, S, Funasaki, N, Iizuka, T, Shiro, Y. | | Deposit date: | 1993-12-24 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural analysis of the myoglobin reconstituted with iron porphine.
J.Biol.Chem., 268, 1993
|
|
1XKV
 
 | | Crystal Structure Of ATP-Dependent Phosphoenolpyruvate Carboxykinase From Thermus thermophilus HB8 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent phosphoenolpyruvate carboxykinase, CALCIUM ION, ... | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-09-29 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
6MU5
 
 | | Bst DNA polymerase I TNA/DNA binary complex | | Descriptor: | DNA (5'-D(P*GP*CP*GP*AP*TP*CP*AP*CP*GP*T)-3'), DNA polymerase I, SULFATE ION, ... | | Authors: | Jackson, L.N, Chim, N, Chaput, J.C. | | Deposit date: | 2018-10-22 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.912 Å) | | Cite: | Crystal structures of a natural DNA polymerase that functions as an XNA reverse transcriptase.
Nucleic Acids Res., 47, 2019
|
|
6MLQ
 
 | | Cryo-EM structure of microtubule-bound Kif7 in the ADP state | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Mani, N, Jiang, S, Wilson-Kubalek, E.M, Ku, P, Milligan, R.A, Subramanian, R. | | Deposit date: | 2018-09-27 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Interplay between the Kinesin and Tubulin Mechanochemical Cycles Underlies Microtubule Tip Tracking by the Non-motile Ciliary Kinesin Kif7.
Dev.Cell, 49, 2019
|
|
6MV6
 
 | | Crystal structure of RNAse 6 | | Descriptor: | PHOSPHATE ION, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
6N7G
 
 | | Cryo-EM structure of tetrameric Ptch1 in complex with ShhNp (form I) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (6.8 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
6MYX
 
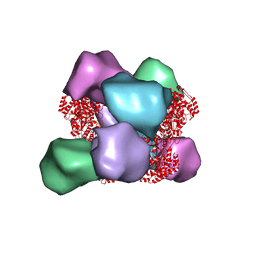 | | EM structure of Bacillus subtilis ribonucleotide reductase inhibited double-helical filament of NrdE alpha subunit with dATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Ribonucleoside-diphosphate reductase | | Authors: | Thomas, W.C, Bacik, J.P, Chen, J.Z, Ando, N. | | Deposit date: | 2018-11-02 | | Release date: | 2019-06-19 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MON
 
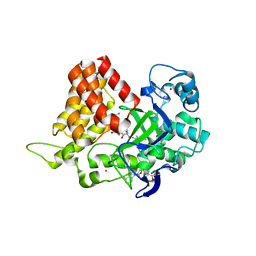 | | Crystal structure of human SMYD2 in complex with Nle-peptide inhibitor | | Descriptor: | GLYCEROL, LYS-LEU-NLE-SER-LYS-ARG-GLY, N-lysine methyltransferase SMYD2, ... | | Authors: | Spellmon, N, Cornett, E, Brunzelle, J, Rothbart, S, Yang, Z. | | Deposit date: | 2018-10-04 | | Release date: | 2018-12-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.711 Å) | | Cite: | A functional proteomics platform to reveal the sequence determinants of lysine methyltransferase substrate selectivity.
Sci Adv, 4, 2018
|
|
6YCF
 
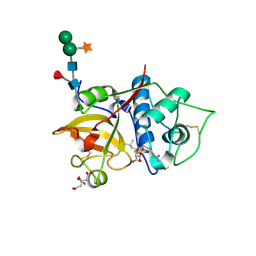 | | Structure the bromelain protease from Ananas comosus in complex with the E64 inhibitor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CITRIC ACID, FBSB, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6N3I
 
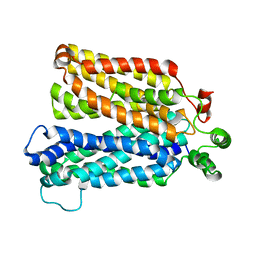 | |
6N4T
 
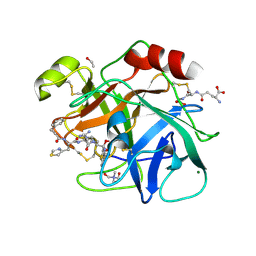 | |
6MUS
 
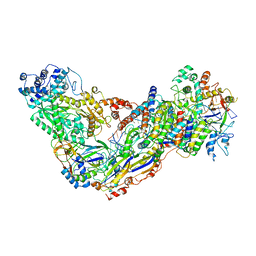 | | Cryo-EM structure of larger Csm-crRNA-target RNA ternary complex in type III-A CRISPR-Cas system | | Descriptor: | RNA (25-MER), RNA (33-MER), Uncharacterized protein Csm1, ... | | Authors: | Jia, N, Wang, C, Eng, E.T. | | Deposit date: | 2018-10-23 | | Release date: | 2018-12-12 | | Last modified: | 2019-01-30 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Type III-A CRISPR-Cas Csm Complexes: Assembly, Periodic RNA Cleavage, DNase Activity Regulation, and Autoimmunity.
Mol. Cell, 73, 2019
|
|
6MV7
 
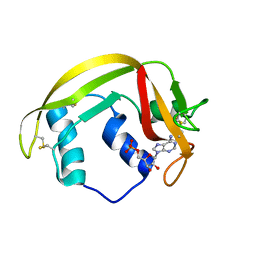 | | Crystal structure of RNAse 6 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Ribonuclease K6 | | Authors: | Couture, J.-F, Doucet, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Insights into Structural and Dynamical Changes Experienced by Human RNase 6 upon Ligand Binding.
Biochemistry, 59, 2020
|
|
6N7K
 
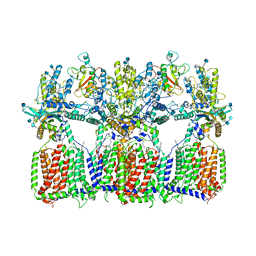 | | Cryo-EM structure of tetrameric Ptch1 in complex with ShhNp (form II) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Yan, N, Gong, X, Qian, H.W. | | Deposit date: | 2018-11-27 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (6.5 Å) | | Cite: | Inhibition of tetrameric Patched1 by Sonic Hedgehog through an asymmetric paradigm.
Nat Commun, 10, 2019
|
|
6N8Z
 
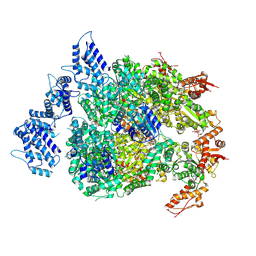 | | HSP104DWB extended conformation | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Heat shock protein 104 | | Authors: | Lee, S, Rho, S.H, Lee, J, Sung, N, Liu, J, Tsai, F.T.F. | | Deposit date: | 2018-11-30 | | Release date: | 2019-01-02 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (9.3 Å) | | Cite: | Cryo-EM Structures of the Hsp104 Protein Disaggregase Captured in the ATP Conformation.
Cell Rep, 26, 2019
|
|
6N20
 
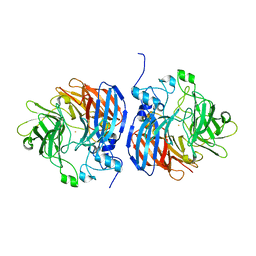 | | Structure of L509V CAO1 - growth condition 2 | | Descriptor: | CHLORIDE ION, Carotenoid oxygenase, FE (II) ION | | Authors: | Khadka, N, Shi, W, Kiser, P.D. | | Deposit date: | 2018-11-12 | | Release date: | 2019-06-05 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Evidence for distinct rate-limiting steps in the cleavage of alkenes by carotenoid cleavage dioxygenases.
J.Biol.Chem., 294, 2019
|
|
6MUA
 
 | |
6NFV
 
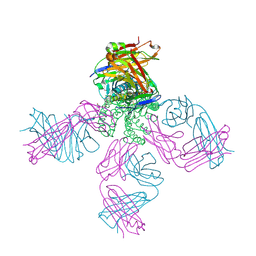 | | Structure of the KcsA-G77C mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
7CK5
 
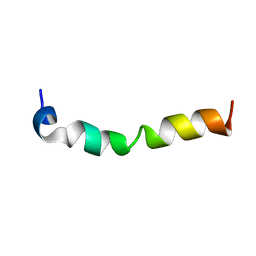 | | Solution structure of 28 amino acid polypeptide (354-381) in Plantago asiatica mosaic virus replicase bound to SDS micelle | | Descriptor: | PlAMV replicase peptide from RNA-dependent RNA polymerase | | Authors: | Komatsu, K, Sasaki, N, Yoshida, T, Suzuki, K, Masujima, Y, Hashimoto, M, Watanabe, S, Tochio, N, Kigawa, T, Yamaji, Y, Oshima, K, Namba, S, Nelson, R, Arie, T. | | Deposit date: | 2020-07-15 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Identification of a Proline-Kinked Amphipathic alpha-Helix Downstream from the Methyltransferase Domain of a Potexvirus Replicase and Its Role in Virus Replication and Perinuclear Complex Formation.
J.Virol., 95, 2021
|
|
6NKG
 
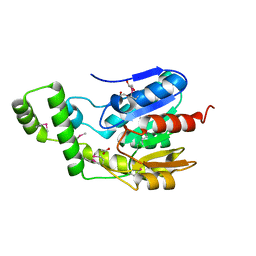 | | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome. | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Lip_vut5, ... | | Authors: | Kim, Y, Welk, L, Mukendi, G, Nkhi, G, Motloi, T, Jedrzejczak, R, Feto, N, Joachimiak, A. | | Deposit date: | 2019-01-07 | | Release date: | 2020-01-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of the Lipase Lip_vut5 from Goat Rumen metagenome.
To Be Published
|
|
6YCG
 
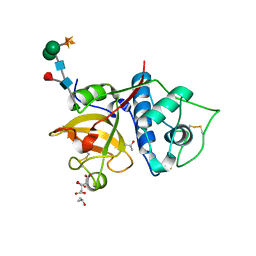 | | Structure the bromelain protease from Ananas comosus in complex with the TLCK inhibitor | | Descriptor: | CITRIC ACID, FBSB, ISOPROPYL ALCOHOL, ... | | Authors: | Azarkan, M, Charlier, P, Herman, R, Delbrassine, F, Sauvage, E, M Rabet, N, Calvo Esposito, R, Kerff, F. | | Deposit date: | 2020-03-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structures of the free and inhibitors-bound forms of bromelain and ananain from Ananas comosus stem and in vitro study of their cytotoxicity.
Sci Rep, 10, 2020
|
|
6NIO
 
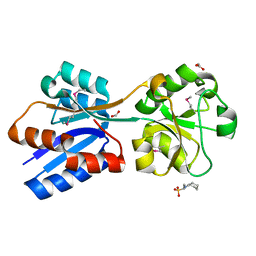 | | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, ACETIC ACID, FORMIC ACID, ... | | Authors: | Kim, Y, Joachimiak, G, Maltseva, N, Endres, M, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-12-31 | | Release date: | 2019-01-16 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Crystal Structure of the Molybdate Transporter Periplasmic Protein ModA from Yersinia pestis
To Be Published
|
|
