6QSX
 
 | | Complement factor B protease domain in complex with the reversible inhibitor ((2S,4S)-1-((5,7-dimethyl-1H-indol-4-yl)methyl)-4-methoxypiperidin-2-yl)methanol. | | Descriptor: | Complement factor B, SULFATE ION, ZINC ION, ... | | Authors: | Adams, C.M, Sellner, H, Ehara, T, Mac Sweeney, A, Crowley, M, Anderson, K, Karki, R, Mainolfi, N, Valeur, E, Sirockin, F, Gerhartz, B, Erbel, P, Hughes, N, Smith, T.M, Cumin, F, Argikar, U, Mogi, M, Sedrani, R, Wiesmann, C, Jaffee, B, Maibaum, J, Flohr, S, Harrison, R, Eder, J. | | Deposit date: | 2019-02-22 | | Release date: | 2019-03-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Small-molecule factor B inhibitor for the treatment of complement-mediated diseases.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2QDQ
 
 | | Crystal structure of the talin dimerisation domain | | Descriptor: | Talin-1 | | Authors: | Gingras, A.R, Putz, N.S.M, Bate, N, Barsukov, I.L, Critchley, D.R.C. | | Deposit date: | 2007-06-21 | | Release date: | 2008-01-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the C-terminal actin-binding domain of talin.
Embo J., 27, 2008
|
|
4YBG
 
 | | Crystal structure of the MAEL domain of Drosophila melanogaster Maelstrom | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Protein maelstrom, ... | | Authors: | Matsumoto, N, Ishitani, R, Nishimasu, H, Nureki, O. | | Deposit date: | 2015-02-18 | | Release date: | 2015-04-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | Crystal Structure and Activity of the Endoribonuclease Domain of the piRNA Pathway Factor Maelstrom
Cell Rep, 11, 2015
|
|
5XXN
 
 | | Crystal Structure of mutant (D286N) beta-glucosidase from Bacteroides thetaiotaomicron in complex with sophorose | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, Periplasmic beta-glucosidase, ... | | Authors: | Nakajima, M, Ishiguro, R, Tanaka, N, Abe, K, Maeda, T, Miyanaga, A, Takahashi, Y, Sugimono, N, Nakai, H, Taguchi, H. | | Deposit date: | 2017-07-04 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Function and structure relationships of a beta-1,2-glucooligosaccharide-degrading beta-glucosidase.
FEBS Lett., 591, 2017
|
|
6F0H
 
 | | Crystal structure ASF1-ip4 | | Descriptor: | CITRIC ACID, GLYCEROL, Histone chaperone ASF1A, ... | | Authors: | Bakail, M, Richet, N, Le Du, M.H, Andreani, J, Guerois, R, Ochsenbein, F. | | Deposit date: | 2017-11-20 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Design on a Rational Basis of High-Affinity Peptides Inhibiting the Histone Chaperone ASF1.
Cell Chem Biol, 26, 2019
|
|
8ALO
 
 | | Heterodimer formation of sensory domains of Vibrio cholerae regulators ToxR and ToxS | | Descriptor: | Cholera toxin transcriptional activator, Transmembrane regulatory protein ToxS | | Authors: | Gubensaek, N, Sagmeister, T, Pavkov-Keller, T, Zangger, K, Buhlheller, C, Wagner, G.E. | | Deposit date: | 2022-08-01 | | Release date: | 2023-09-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.002 Å) | | Cite: | Vibrio cholerae's ToxRS bile sensing system.
Elife, 12, 2023
|
|
7Y1Q
 
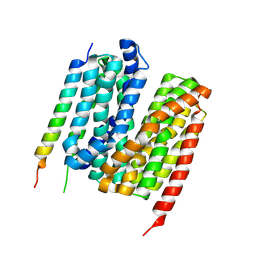 | | 5.0 angstrom cryo-EM structure of transmembrane regions of mouse Basigin/MCT1 in complex with antibody 6E7F1 | | Descriptor: | Isoform 2 of Basigin, Monocarboxylate transporter 1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (5.03 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin-2
To Be Published
|
|
7Y1B
 
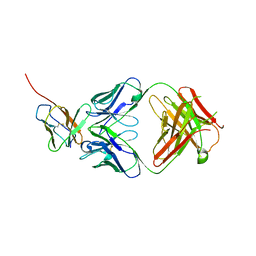 | | 3.2 angstrom cryo-EM structure of extracellular region of mouse Basigin-2 in complex with the Fab fragment of antibody 6E7F1 | | Descriptor: | Heavy chain of 6E7F1, Isoform 2 of Basigin, Light chain of 6E7F1 | | Authors: | Zhang, H, Yang, X, Xue, Y, Huang, Y, Mo, X, Zhang, H, Li, N, Gao, N, Li, X, Wang, S, Gao, Y, Liao, J. | | Deposit date: | 2022-06-08 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.23 Å) | | Cite: | Allosteric modulation of monocarboxylate transporters 1 and 4 by targeting their chaperon Basigin
To Be Published
|
|
1ILZ
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 6.1 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
6F1F
 
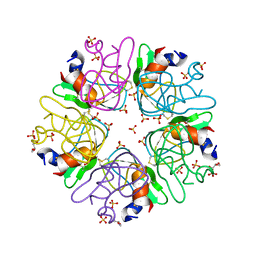 | | The methylene thioacetal BPTI (Bovine Pancreatic Trypsin Inhibitor) mutant structure | | Descriptor: | GLYCEROL, Pancreatic trypsin inhibitor, SULFATE ION | | Authors: | Lansky, S, Mousa, R, Metanis, N, Shoham, G. | | Deposit date: | 2017-11-21 | | Release date: | 2018-07-18 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.716 Å) | | Cite: | BPTI folding revisited: switching a disulfide into methylene thioacetal reveals a previously hidden path.
Chem Sci, 9, 2018
|
|
5MHF
 
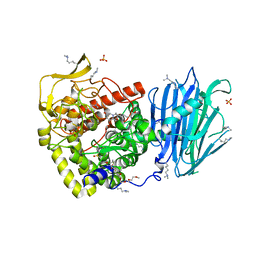 | | Murine endoplasmic reticulum alpha-glucosidase I with N-9'-methoxynonyl-1-deoxynojirimycin. | | Descriptor: | 2,5,8,11,14,17,20,23-OCTAOXAPENTACOSAN-25-OL, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mannosyl-oligosaccharide glucosidase, ... | | Authors: | Hill, J.C, Caputo, A.T, Roversi, P, Zitzmann, N. | | Deposit date: | 2016-11-24 | | Release date: | 2017-12-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting Endoplasmic Reticulum alpha-Glucosidase I with a Single-Dose Iminosugar Treatment Protects against Lethal Influenza and Dengue Virus Infections.
J.Med.Chem., 2020
|
|
4C7D
 
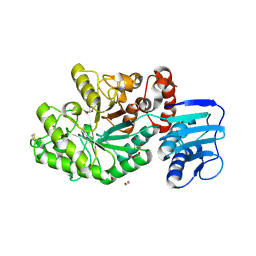 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
1WP4
 
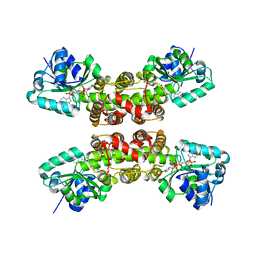 | |
4C7G
 
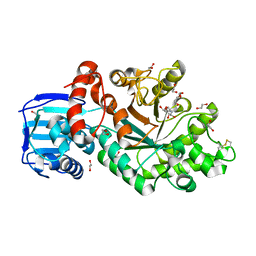 | | Structure and activity of the GH20 beta-N-acetylhexosaminidase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, 2-METHYL-4,5-DIHYDRO-(1,2-DIDEOXY-ALPHA-D-GLUCOPYRANOSO)[2,1-D]-1,3-OXAZOLE, BETA-N-ACETYLHEXOSAMINIDASE | | Authors: | Nguyenthi, N, Offen, W.A, Davies, G.J, Doucet, N. | | Deposit date: | 2013-09-20 | | Release date: | 2014-03-12 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure and Activity of the Streptomyces Coelicolor A3(2) Beta-N-Acetylhexosaminidase Provides Further Insight Into Gh20 Family Catalysis and Inhibition.
Biochemistry, 53, 2014
|
|
4Z89
 
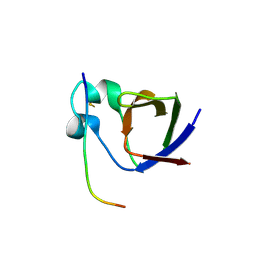 | | SH3-II of Drosophila Rim-binding protein bound to a Cacophony derived peptide | | Descriptor: | CALCIUM ION, RIM-binding protein, isoform F, ... | | Authors: | Driller, J.H, Holton, N, Siebert, M, Boehme, M.A, Wahl, M.C, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-04-08 | | Release date: | 2015-08-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | A high affinity RIM-binding protein/Aplip1 interaction prevents the formation of ectopic axonal active zones.
Elife, 4, 2015
|
|
1J1W
 
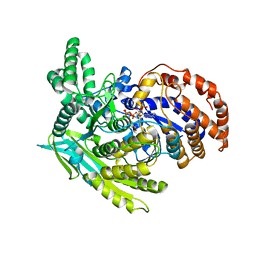 | | Crystal Structure Of The Monomeric Isocitrate Dehydrogenase In Complex With NADP+ | | Descriptor: | Isocitrate Dehydrogenase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-12-19 | | Release date: | 2003-09-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal Structure of the Monomeric Isocitrate Dehydrogenase in the Presence of NADP+
J.Biol.Chem., 278, 2003
|
|
1J4J
 
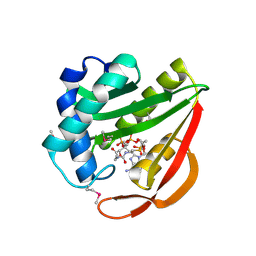 | | Crystal Structure of Tabtoxin Resistance Protein (form II) complexed with an Acyl Coenzyme A | | Descriptor: | ACETYL COENZYME *A, TABTOXIN RESISTANCE PROTEIN | | Authors: | He, H, Ding, Y, Bartlam, M, Zhang, R, Duke, N, Joachimiak, A, Shao, Y, Cao, Z, Tang, H, Liu, Y, Jiang, F, Liu, J, Zhao, N, Rao, Z. | | Deposit date: | 2001-10-02 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of tabtoxin resistance protein complexed with acetyl coenzyme A reveals the mechanism for beta-lactam acetylation.
J.Mol.Biol., 325, 2003
|
|
5MWU
 
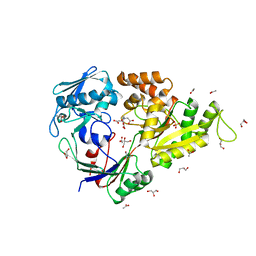 | | Crystal structure of the periplasmic nickel-binding protein NikA from Escherichia coli in complex with Ru(bpza)(CO)2Cl | | Descriptor: | ACETATE ION, CARBON MONOXIDE, CHLORIDE ION, ... | | Authors: | Cavazza, C, Lopez, S, Rondot, L, Iannello, M, Boeri-Erba, E, Burzlaff, N, Strinitz, F, Jorge-Robin, A, Marchi-Delapierre, C, Menage, S. | | Deposit date: | 2017-01-20 | | Release date: | 2017-02-22 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Efficient conversion of alkenes to chlorohydrins by a Ru-based artificial enzyme
To Be Published
|
|
8BN6
 
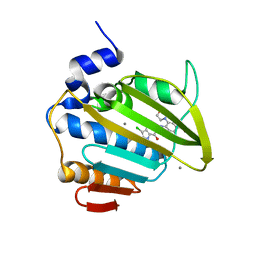 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with EBL3021 | | Descriptor: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-morpholin-4-yl-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, DNA gyrase subunit B | | Authors: | Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | Deposit date: | 2022-11-12 | | Release date: | 2023-03-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | New Dual Inhibitors of Bacterial Topoisomerases with Broad-Spectrum Antibacterial Activity and In Vivo Efficacy against Vancomycin-Intermediate Staphylococcus aureus .
J.Med.Chem., 66, 2023
|
|
1ILD
 
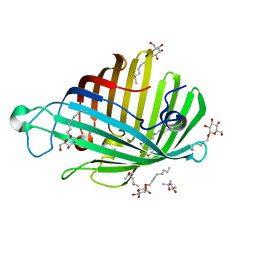 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT pH 4.6 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHOSPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-08 | | Release date: | 2001-10-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
4ZNQ
 
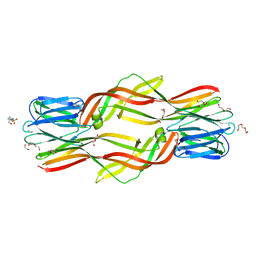 | | Crystal structure of Dln1 complexed with Man(alpha1-2)Man | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Jia, N, Jiang, Y.L, Cheng, W, Wang, H.W, Zhou, C.Z, Chen, Y. | | Deposit date: | 2015-05-05 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for receptor recognition and pore formation of a zebrafish aerolysin-like protein.
Embo Rep., 17, 2016
|
|
1IM0
 
 | | OUTER MEMBRANE PHOSPHOLIPASE A FROM ESCHERICHIA COLI N156A ACTIVE SITE MUTANT PH 8.3 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, OUTER MEMBRANE PHSOPHOLIPASE A, octyl beta-D-glucopyranoside | | Authors: | Snijder, H.J, Van Eerde, J.H, Kingma, R.L, Kalk, K.H, Dekker, N, Egmond, M.R, Dijkstra, B.W. | | Deposit date: | 2001-05-09 | | Release date: | 2001-10-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.98 Å) | | Cite: | Structural investigations of the active-site mutant Asn156Ala of outer membrane phospholipase A: function of the Asn-His interaction in the catalytic triad.
Protein Sci., 10, 2001
|
|
2RNE
 
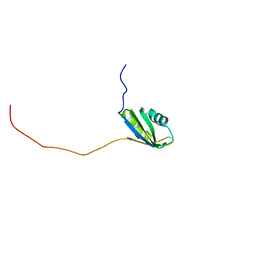 | | Solution structure of the second RNA recognition motif (RRM) of TIA-1 | | Descriptor: | Tia1 protein | | Authors: | Takahashi, M, Kuwasako, K, Abe, C, Tsuda, K, Inoue, M, Terada, T, Shirouzu, M, Kobayashi, N, Kigawa, T, Taguchi, S, Guntert, P, Hayashizaki, Y, Tanaka, A, Muto, Y, Yokoyama, S. | | Deposit date: | 2007-12-19 | | Release date: | 2008-11-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the second RNA recognition motif (RRM) domain of murine T cell intracellular antigen-1 (TIA-1) and its RNA recognition mode
Biochemistry, 47, 2008
|
|
5M8I
 
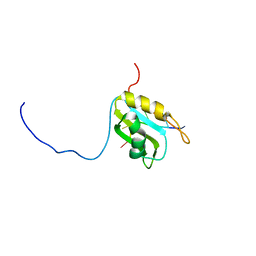 | |
1X1K
 
 | | Host-guest peptide (Pro-Pro-Gly)4-(Pro-alloHyp-Gly)-(Pro-Pro-Gly)4 | | Descriptor: | Host-guest peptide (Pro-Pro-Gly)4-(Pro-alloHyp-Gly)-(Pro-Pro-Gly)4 | | Authors: | Jiravanichanun, N, Hongo, C, Wu, G, Noguchi, K, Okuyama, K, Nishino, N, Silva, T. | | Deposit date: | 2005-04-05 | | Release date: | 2005-06-14 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Unexpected puckering of hydroxyproline in the guest triplets, hyp-pro-gly and pro-allohyp-gly sandwiched between pro-pro-gly sequence
Chembiochem, 6, 2005
|
|
