5A9Q
 
 | | Human nuclear pore complex | | Descriptor: | NUCLEAR PORE COMPLEX PROTEIN NUP107, NUCLEAR PORE COMPLEX PROTEIN NUP133, NUCLEAR PORE COMPLEX PROTEIN NUP155, ... | | Authors: | von Appen, A, Kosinski, J, Sparks, L, Ori, A, DiGuilio, A, Vollmer, B, Mackmull, M, Banterle, N, Parca, L, Kastritis, P, Buczak, K, Mosalaganti, S, Hagen, W, Andres-Pons, A, Lemke, E.A, Bork, P, Antonin, W, Glavy, J.S, Bui, K.H, Beck, M. | | Deposit date: | 2015-07-22 | | Release date: | 2015-09-30 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | In Situ Structural Analysis of the Human Nuclear Pore Complex
Nature, 526, 2015
|
|
6UE7
 
 | | Structure of dimeric sIgA complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin J chain, ... | | Authors: | Kumar, N, Arthur, C.P, Ciferri, C, Matsumoto, M.L. | | Deposit date: | 2019-09-20 | | Release date: | 2020-02-19 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the secretory immunoglobulin A core.
Science, 367, 2020
|
|
4ZY1
 
 | |
6T59
 
 | | Structure of rabbit 80S ribosome translating beta-tubulin in complex with tetratricopeptide protein 5 and nascent chain-associated complex | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Lin, Z, Gasic, I, Chandrasekaran, V, Peters, N, Shao, S, Ramakrishnan, V, Mitchison, T.J, Hegde, R.S. | | Deposit date: | 2019-10-15 | | Release date: | 2019-11-27 | | Last modified: | 2020-01-15 | | Method: | ELECTRON MICROSCOPY (3.11 Å) | | Cite: | TTC5 mediates autoregulation of tubulin via mRNA degradation.
Science, 367, 2020
|
|
5A2I
 
 | | Crystal structure of scFv-SM3 in complex with APD-SGalNAc-RP | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-alpha-D-galactopyranose, ANTIGEN TN, ... | | Authors: | Martinez-Saez, N, Castro-Lopez, J, Valero-Gonzalez, J, Madariaga, D, Companon, I, Somovilla, V.J, Salvado, M, Asensio, J.L, Jimenez-Barbero, J, Avenoza, A, Busto, J.H, Bernardes, G.J.L, Peregrina, J.M, Hurtado-Guerrero, R, Corzana, F. | | Deposit date: | 2015-05-20 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Deciphering the Non-Equivalence of Serine and Threonine O-Glycosylation Points: Implications for Molecular Recognition of the Tn Antigen by an Anti-Muc1 Antibody.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5ABF
 
 | | Structure of GH84 with ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2S,3R,4R,5R)-5-(hydroxymethyl)-3,4-bis(oxidanyl)-1-pentyl-pyrrolidin-2-yl]-N-methyl-ethanamide, CALCIUM ION, ... | | Authors: | Bergeron-Brlek, M, Goodwin-Tindall, J, Cekic, N, Varghese, V, Zandberg, W.F, Shan, X, Roth, C, Chan, S, Davies, G.J, Vocadlo, D.J, Britton, R. | | Deposit date: | 2015-08-05 | | Release date: | 2015-11-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Convenient Approach to Stereoisomeric Iminocyclitols: Generation of Potent Brain-Permeable Oga Inhibitors.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
5A3M
 
 | | Structure of Cea1A in complex with Chitobiose | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CEA1, ... | | Authors: | Kock, M, Brueckner, S, Wozniak, N, Veelders, M, Schlereth, J, Moesch, H.-U, Essen, L.-O. | | Deposit date: | 2015-06-02 | | Release date: | 2016-06-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | High-Affinity Recognition of Non-Reducing Chitinous Ends by the Yeast Adhesin Cea1
To be Published
|
|
5A3F
 
 | | Crystal structure of the dynamin tetramer | | Descriptor: | DYNAMIN 3 | | Authors: | Reubold, T.F, Faelber, K, Plattner, N, Posor, Y, Branz, K, Curth, U, Schlegel, J, Anand, R, Manstein, D.J, Noe, F, Haucke, V, Daumke, O, Eschenburg, S. | | Deposit date: | 2015-05-29 | | Release date: | 2015-08-26 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Crystal Structure of the Dynamin Tetramer
Nature, 525, 2015
|
|
4ZZB
 
 | | The GLIC pentameric Ligand-Gated Ion Channel Locally-closed form complexed to xenon | | Descriptor: | ACETATE ION, CHLORIDE ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sauguet, L, Fourati, Z, Prange, T, Delarue, M, Colloc'h, N. | | Deposit date: | 2015-05-22 | | Release date: | 2016-03-02 | | Last modified: | 2018-11-21 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural Basis for Xenon Inhibition in a Cationic Pentameric Ligand-Gated Ion Channel.
Plos One, 11, 2016
|
|
5A16
 
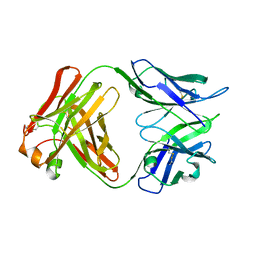 | | Crystal structure of Fab4201 raised against Human Erythrocyte Anion Exchanger 1 | | Descriptor: | FAB4201 HEAVY CHAIN | | Authors: | Arakawa, T, Kobayashi-Yugiri, T, Alguel, Y, Weyand, S, Iwanari, H, Hatae, H, Iwata, M, Abe, Y, Hino, T, Ikeda-Suno, C, Kuma, H, Kang, D, Murata, T, Hamakubo, T, Cameron, A, Kobayashi, T, Hamasaki, N, Iwata, S. | | Deposit date: | 2015-04-28 | | Release date: | 2015-06-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Anion Exchanger Domain of Human Erythrocyte Band 3
Science, 350, 2015
|
|
5A22
 
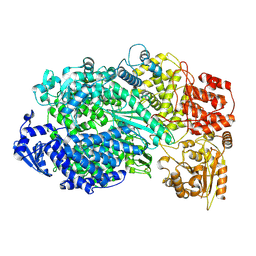 | | Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy | | Descriptor: | VESICULAR STOMATITIS VIRUS L POLYMERASE, ZINC ION | | Authors: | Liang, B, Li, Z, Jenni, S, Rameh, A.A, Morin, B.M, Grant, T, Grigorieff, N, Harrison, S.C, Whelan, S.P.J. | | Deposit date: | 2015-05-06 | | Release date: | 2015-08-19 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure of the L Protein of Vesicular Stomatitis Virus from Electron Cryomicroscopy.
Cell(Cambridge,Mass.), 162, 2015
|
|
5A7Q
 
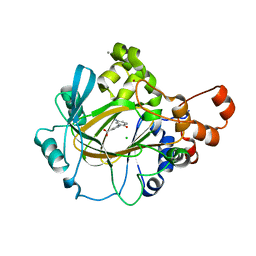 | | Crystal structure of human JMJD2A in complex with compound 30 | | Descriptor: | 1,2-ETHANEDIOL, 2-(5-azanyl-2-oxidanyl-phenyl)pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Dixon-Clarke, S, MacKenzie, A, Nowak, R, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
6TMX
 
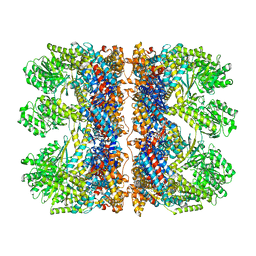 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ATPgammaS | | Descriptor: | MAGNESIUM ION, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
5AKC
 
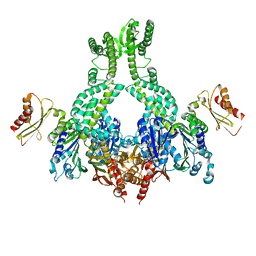 | | MutS in complex with the N-terminal domain of MutL - crystal form 2 | | Descriptor: | DNA MISMATCH REPAIR PROTEIN MUTL, DNA MISMATCH REPAIR PROTEIN MUTS, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Groothuizen, F.S, Winkler, I, Cristovao, M, Fish, A, Winterwerp, H.H.K, Reumer, A, Marx, A.D, Hermans, N, Nicholls, R.A, Murshudov, G.N, Lebbink, J.H.G, Friedhoff, P, Sixma, T.K. | | Deposit date: | 2015-03-03 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (6.6 Å) | | Cite: | MutS/MutL crystal structure reveals that the MutS sliding clamp loads MutL onto DNA.
Elife, 4, 2015
|
|
2AHJ
 
 | | NITRILE HYDRATASE COMPLEXED WITH NITRIC OXIDE | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, FE (III) ION, NITRIC OXIDE, ... | | Authors: | Nagashima, S, Nakasako, M, Dohmae, N, Tsujimura, M, Takio, K, Odaka, M, Yohda, M, Kamiya, N, Endo, I. | | Deposit date: | 1997-12-24 | | Release date: | 1999-01-27 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel non-heme iron center of nitrile hydratase with a claw setting of oxygen atoms.
Nat.Struct.Biol., 5, 1998
|
|
6TM8
 
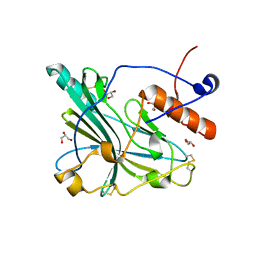 | | Crystal structure of glycoprotein D of Equine Herpesvirus Type 4 | | Descriptor: | Envelope glycoprotein D, GLYCEROL | | Authors: | Kremling, V, Loll, B, Osterrieder, N, Wahl, M, Dahmani, I, Chiantia, P, Azab, W. | | Deposit date: | 2019-12-03 | | Release date: | 2020-11-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of glycoprotein D of equine alphaherpesviruses reveal potential binding sites to the entry receptor MHC-I.
Front Microbiol, 14, 2023
|
|
6TMQ
 
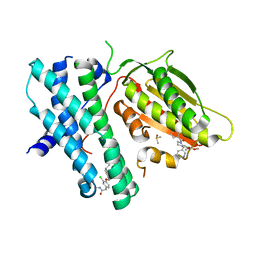 | | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures | | Descriptor: | DIMETHYL SULFOXIDE, N-(2-AMINOETHYL)-2-{3-CHLORO-4-[(4-ISOPROPYLBENZYL)OXY]PHENYL} ACETAMIDE, [2,4-bis(oxidanyl)phenyl]-[(1~{S})-6,7-dimethoxy-1-pyridin-3-yl-3,4-dihydro-1~{H}-isoquinolin-2-yl]methanone, ... | | Authors: | Baker, L.M, Aimon, A, Murray, J.B, Surgenor, A.E, Matassova, N, Roughley, S.D, von Delft, F, Hubbard, R.E. | | Deposit date: | 2019-12-05 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.112 Å) | | Cite: | Rapid optimisation of fragments and hits to lead compounds from screening of crude reaction mixtures
Commun Chem, 2020
|
|
6TMW
 
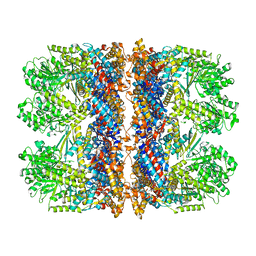 | | Structure of the chaperonin gp146 from the bacteriophage EL (Pseudomonas aeruginosa) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Putative GroEL-like chaperonine protein | | Authors: | Bracher, A, Wang, H, Paul, S.S, Wischnewski, N, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2019-12-05 | | Release date: | 2020-04-22 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (5.91 Å) | | Cite: | Structure and conformational cycle of a bacteriophage-encoded chaperonin.
Plos One, 15, 2020
|
|
5AFS
 
 | | structure of Zn-bound periplasmic metal binding protein from candidatus liberibacter asiaticus | | Descriptor: | ACETATE ION, GLYCEROL, PERIPLASMIC SOLUTE BINDING PROTEIN, ... | | Authors: | Sharma, N, Selvakumar, P, Kumar, P, Sharma, A.K. | | Deposit date: | 2015-01-23 | | Release date: | 2016-02-24 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure analysis in Zn(2+)-bound state and biophysical characterization of CLas-ZnuA2.
Biochim. Biophys. Acta, 1864, 2016
|
|
5A7P
 
 | | Crystal structure of human JMJD2A in complex with compound 36 | | Descriptor: | 1,2-ETHANEDIOL, 2-[5-[(5-methyl-1,2-oxazol-3-yl)carbonylamino]-2-oxidanyl-phenyl]pyridine-4-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Nowak, R, Velupillai, S, Krojer, T, Gileadi, C, Johansson, C, Korczynska, M, Le, D.D, Younger, N, Gregori-Puigjane, E, Tumber, A, Iwasa, E, Pollock, S.B, Ortiz Torres, I, Kopec, J, Tallant, C, Froese, S, von Delft, F, Arrowsmith, C.H, Bountra, C, Edwards, A, Shoichet, B.K, Fujimori, D.G, Oppermann, U. | | Deposit date: | 2015-07-09 | | Release date: | 2016-01-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Docking and Linking of Fragments to Discover Jumonji Histone Demethylase Inhibitors.
J.Med.Chem., 59, 2016
|
|
6TQN
 
 | | rrn anti-termination complex without S4 | | Descriptor: | 30S ribosomal protein S10, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Huang, Y.H, Wahl, M.C, Loll, B, Hilal, T, Said, N. | | Deposit date: | 2019-12-17 | | Release date: | 2020-08-05 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structure-Based Mechanisms of a Molecular RNA Polymerase/Chaperone Machine Required for Ribosome Biosynthesis.
Mol.Cell, 79, 2020
|
|
5AIJ
 
 | | P. aeruginosa SdsA hexagonal polymorph | | Descriptor: | ALKYL SULFATASE, GLYCEROL, ZINC ION | | Authors: | De la Mora, E, Flores-Hernandez, E, Jakoncic, J, Stojanoff, V, Sanchez-Puig, N, Moreno, A. | | Deposit date: | 2015-02-13 | | Release date: | 2015-10-07 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Sdsa Polymorph Isolation and Improvement of Their Crystal Quality Using Nonconventional Crystallization Techniques
J.Appl.Crystallogr., 48, 2015
|
|
5A77
 
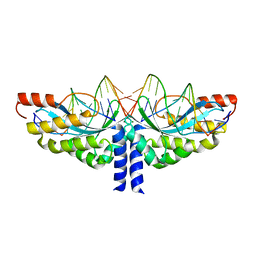 | | Crystal structure of the homing endonuclease I-CvuI in complex with I- CreI target (C1221) in the presence of 2 mM Mg revealing DNA cleaved | | Descriptor: | 10MER DNA, 5'-D(*GP*AP*CP*GP*TP*TP*TP*TP* GP*AP*DGP*AP*CP*GP*TP*TP*TP*TP*GP*A)-3', 14MER DNA, ... | | Authors: | Molina, R, Redondo, P, LopezMendez, B, Villate, M, Merino, N, Blanco, F.J, Valton, J, Grizot, S, Duchateau, P, Prieto, J, Montoya, G. | | Deposit date: | 2015-07-03 | | Release date: | 2015-09-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of the Homing Endonuclease I-Cvui Provides a New Template for Genome Modification
J.Biol.Chem., 290, 2015
|
|
4ZX3
 
 | |
5A3V
 
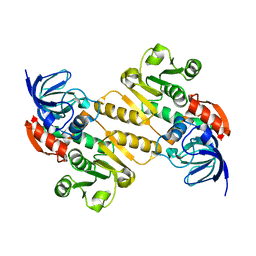 | | Crystal structure of the chloroplastic gamma-ketol reductase from Arabidopsis thaliana | | Descriptor: | PUTATIVE QUINONE-OXIDOREDUCTASE HOMOLOG, CHLOROPLASTIC | | Authors: | Mas-y-mas, S, Curien, G, Giustini, C, Rolland, N, Ferrer, J.L, Cobessi, D. | | Deposit date: | 2015-06-03 | | Release date: | 2016-09-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of the Chloroplastic Oxoene Reductase ceQORH from Arabidopsis thaliana.
Front Plant Sci, 8, 2017
|
|
