2ZXM
 
 | | A New Class of Vitamin D Receptor Ligands that Induce Structural Rearrangement of the Ligand-binding Pocket | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-[(1R,3aS,7aR)-1-[(2R,3S)-3-(2-hydroxyethyl)heptan-2-yl]-7a-methyl-2,3,3a,5,6,7-hexahydro-1H-inden-4-ylidene]ethylidene]-4-methylidene-cyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Nakabayashi, M, Ikura, T, Ito, N. | | Deposit date: | 2009-01-04 | | Release date: | 2009-02-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | A New Class of Vitamin D Analogues that Induce Structural Rearrangement of the Ligand-Binding Pocket of the Receptor
J.Med.Chem., 52, 2009
|
|
1BZD
 
 | |
1C0G
 
 | | CRYSTAL STRUCTURE OF 1:1 COMPLEX BETWEEN GELSOLIN SEGMENT 1 AND A DICTYOSTELIUM/TETRAHYMENA CHIMERA ACTIN (MUTANT 228: Q228K/T229A/A230Y/E360H) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, PROTEIN (CHIMERIC ACTIN), ... | | Authors: | Matsuura, Y, Stewart, M, Kawamoto, M, Kamiya, N, Saeki, K, Yasunaga, T, Wakabayashi, T. | | Deposit date: | 1999-07-16 | | Release date: | 2000-03-01 | | Last modified: | 2022-12-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for the higher Ca(2+)-activation of the regulated actin-activated myosin ATPase observed with Dictyostelium/Tetrahymena actin chimeras.
J.Mol.Biol., 296, 2000
|
|
1QQS
 
 | | NEUTROPHIL GELATINASE ASSOCIATED LIPOCALIN HOMODIMER | | Descriptor: | DECANOIC ACID, NEUTROPHIL GELATINASE, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goetz, D.H, Willie, S.T, Armen, R, Bratt, T, Borregaard, N, Strong, R.K. | | Deposit date: | 1999-06-07 | | Release date: | 2000-04-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Ligand preference inferred from the structure of neutrophil gelatinase associated lipocalin
Biochemistry, 39, 2000
|
|
1C3Q
 
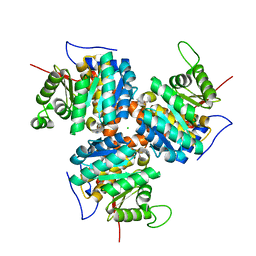 | | CRYSTAL STRUCTURE OF NATIVE THIAZOLE KINASE IN THE MONOCLINIC FORM | | Descriptor: | 2-(4-METHYL-THIAZOL-5-YL)-ETHANOL, CHLORIDE ION, Hydroxyethylthiazole kinase | | Authors: | Campobasso, N, Mathews, I.I, Begley, T.P, Ealick, S.E. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of 4-methyl-5-beta-hydroxyethylthiazole kinase from Bacillus subtilis at 1.5 A resolution.
Biochemistry, 39, 2000
|
|
1QVW
 
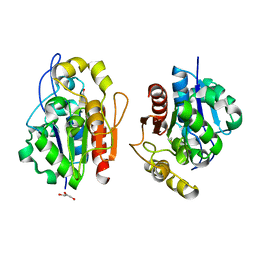 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1T6H
 
 | | Crystal Structure T4 Lysozyme incorporating an unnatural amino acid p-iodo-L-phenylalanine at position 153 | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Lysozyme | | Authors: | Spraggon, G, Xie, J, Wang, L, Wu, N, Brock, A, Schultz, P.G. | | Deposit date: | 2004-05-06 | | Release date: | 2004-10-26 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The site-specific incorporation of p-iodo-L-phenylalanine into proteins for structure determination.
Nat.Biotechnol., 22, 2004
|
|
1TEV
 
 | | Crystal structure of the human UMP/CMP kinase in open conformation | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Segura-Pena, D, Sekulic, N, Ort, S, Konrad, M, Lavie, A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced Conformational Changes in Human UMP/CMP Kinase.
J.Biol.Chem., 279, 2004
|
|
1TGM
 
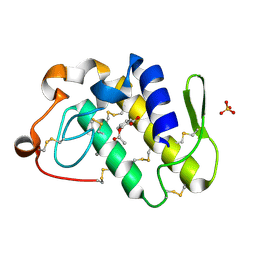 | | Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution | | Descriptor: | 2-(ACETYLOXY)BENZOIC ACID, CALCIUM ION, Phospholipase A2, ... | | Authors: | Singh, N, Jabeen, T, Sharma, S, Bhushan, A, Singh, T.P. | | Deposit date: | 2004-05-28 | | Release date: | 2004-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of a complex formed between group II phospholipase A2 and aspirin at 1.86 A resolution
To be Published
|
|
1TJK
 
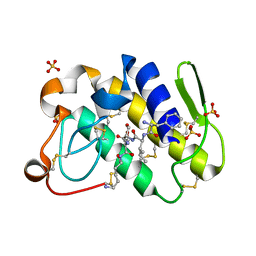 | | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe- Leu- Ser- Thr- Lys at 1.2 A resolution | | Descriptor: | Phospholipase A2, SULFATE ION, synthetic peptide | | Authors: | Singh, N, Jabeen, T, Somvanshi, R.K, Sharma, S, Perbandt, M, Dey, S, Betzel, C, Singh, T.P. | | Deposit date: | 2004-06-06 | | Release date: | 2004-06-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of the complex formed between group II phospholipase A2 with a designed pentapeptide, Phe - Leu - Ser - Thr - Lys at 1.2 A resolution
To be Published
|
|
1TD2
 
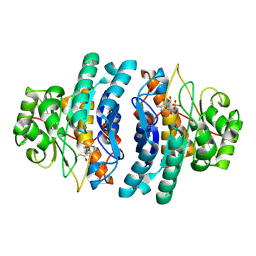 | | Crystal Structure of the PdxY Protein from Escherichia coli | | Descriptor: | 3-HYDROXY-5-(HYDROXYMETHYL)-2-METHYLISONICOTINALDEHYDE, Pyridoxamine kinase, SULFATE ION | | Authors: | Safo, M.K, Musayev, F.N, Hunt, S, di Salvo, M, Scarsdale, N, Schirch, V. | | Deposit date: | 2004-05-21 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Crystal structure of the PdxY Protein from Escherichia coli
J.Bacteriol., 186, 2004
|
|
1T21
 
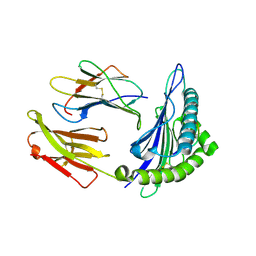 | | Structural basis for degenerate recognition of HIV peptide variants by cytotoxic lymphocyte, variant SL9, monoclinic crystal | | Descriptor: | Beta-2-microglobulin, GAG PEPTIDE, HLA class I histocompatibility antigen, ... | | Authors: | Martinez-Hackert, E, Anikeeva, N, Kalams, S.A, Walker, B.D, Hendrickson, W.A, Sykulev, Y. | | Deposit date: | 2004-04-19 | | Release date: | 2005-09-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Basis for Degenerate Recognition of Natural HIV Peptide Variants by Cytotoxic Lymphocytes.
J.Biol.Chem., 281, 2006
|
|
1SV3
 
 | | Structure of the complex formed between Phospholipase A2 and 4-methoxybenzoic acid at 1.3A resolution. | | Descriptor: | 4-METHOXYBENZOIC ACID, Phospholipase A2, SULFATE ION | | Authors: | Singh, N, Prahathees, E, Jabeen, T, Pal, A, Ethayathulla, A.S, Prem kumar, R, Sharma, S, Singh, T.P. | | Deposit date: | 2004-03-27 | | Release date: | 2004-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structures of the complexes of a group IIA phospholipase A2 with two natural anti-inflammatory agents, anisic acid, and atropine reveal a similar mode of binding
Proteins, 64, 2006
|
|
1CCR
 
 | | STRUCTURE OF RICE FERRICYTOCHROME C AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Ochi, H, Hata, Y, Tanaka, N, Kakudo, M, Sakurai, T, Aihara, S, Morita, Y. | | Deposit date: | 1983-03-14 | | Release date: | 1983-04-21 | | Last modified: | 2021-03-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of rice ferricytochrome c at 2.0 A resolution.
J.Mol.Biol., 166, 1983
|
|
1SYS
 
 | | Crystal structure of HLA, B*4403, and peptide EEPTVIKKY | | Descriptor: | Beta-2-microglobulin, Sorting nexin 5, leukocyte antigen (HLA) class I molecule | | Authors: | Zernich, D, Purcell, A.W, Macdonald, W.A, Kjer-Nielsen, L, Ely, L.K, Laham, N, Crockford, T, Mifsud, N.A, Tait, B.D, Holdsworth, R, Brooks, A.G, Bottomley, S.P, Beddoe, T, Peh, C.A, Rossjohn, J, McCluskey, J. | | Deposit date: | 2004-04-01 | | Release date: | 2004-10-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion
J.Exp.Med., 200, 2004
|
|
1C94
 
 | | REVERSING THE SEQUENCE OF THE GCN4 LEUCINE ZIPPER DOES NOT AFFECT ITS FOLD. | | Descriptor: | RETRO-GCN4 LEUCINE ZIPPER | | Authors: | Mittl, P.R.E, Deillon, C.A, Sargent, D, Liu, N, Klauser, S, Thomas, R.M, Gutte, B, Gruetter, M.G. | | Deposit date: | 1999-07-30 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | The retro-GCN4 leucine zipper sequence forms a stable three-dimensional structure.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1B4B
 
 | | STRUCTURE OF THE OLIGOMERIZATION DOMAIN OF THE ARGININE REPRESSOR FROM BACILLUS STEAROTHERMOPHILUS | | Descriptor: | ARGININE, ARGININE REPRESSOR | | Authors: | Ni, J, Sakanyan, V, Charlier, D, Glansdorff, N, Van Duyne, G.D. | | Deposit date: | 1998-12-18 | | Release date: | 1999-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the arginine repressor from Bacillus stearothermophilus.
Nat.Struct.Biol., 6, 1999
|
|
2Z9Z
 
 | | Crystal structure of CERT START domain(N504A mutant), in complex with C10-diacylglycerol | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl didecanoate, Lipid-transfer protein CERT | | Authors: | Kudo, N, Kumagai, K, Wakatsuki, S, Nishijima, M, Hanada, K, Kato, R. | | Deposit date: | 2007-09-26 | | Release date: | 2008-02-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis for specific lipid recognition by CERT responsible for nonvesicular trafficking of ceramide.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
1B7A
 
 | | STRUCTURE OF THE PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN FROM BOVINE BRAIN | | Descriptor: | PHOSPHATIDYLETHANOLAMINE-BINDING PROTEIN, PHOSPHORIC ACID MONO-(2-AMINO-ETHYL) ESTER | | Authors: | Serre, L, Vallee, B, Bureaud, N, Schoentgen, F, Zelwer, C. | | Deposit date: | 1999-01-21 | | Release date: | 1999-01-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the phosphatidylethanolamine-binding protein from bovine brain: a novel structural class of phospholipid-binding proteins.
Structure, 6, 1998
|
|
1REZ
 
 | | HUMAN LYSOZYME-N-ACETYLLACTOSAMINE COMPLEX | | Descriptor: | GLYCEROL, LYSOZYME, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Muraki, M, Harata, K, Sugita, N, Sato, K. | | Deposit date: | 1996-08-21 | | Release date: | 1997-02-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Origin of carbohydrate recognition specificity of human lysozyme revealed by affinity labeling.
Biochemistry, 35, 1996
|
|
1R7D
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Ensemble of 51 structures, sample in 50% tfe) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
1RU4
 
 | | Crystal structure of pectate lyase Pel9A | | Descriptor: | CALCIUM ION, Pectate lyase | | Authors: | Jenkins, J, Shevchik, V.E, Hugouvieux-Cotte-Pattat, N, Pickersgill, R.W. | | Deposit date: | 2003-12-11 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of pectate lyase Pel9A from Erwinia chrysanthemi
J.Biol.Chem., 279, 2004
|
|
1RVX
 
 | | 1934 H1 Hemagglutinin in complex with LSTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Gamblin, S.J, Haire, L.F, Russell, R.J, Stevens, D.J, Xiao, B, Ha, Y, Vasisht, N, Steinhauer, D.A, Daniels, R.S, Elliot, A, Wiley, D.C, Skehel, J.J. | | Deposit date: | 2003-12-15 | | Release date: | 2004-03-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure and receptor binding properties of the 1918 influenza hemagglutinin.
Science, 303, 2004
|
|
2ZIS
 
 | | Crystal Structure of rat protein farnesyltransferase complexed with a bezoruran inhibitor and FPP | | Descriptor: | 3-{2-[(S)-(4-cyanophenyl)(hydroxy)(1-methyl-1H-imidazol-5-yl)methyl]-5-nitro-1-benzofuran-7-yl}benzonitrile, ACETIC ACID, FARNESYL DIPHOSPHATE, ... | | Authors: | Fukami, T.A, Sogabe, S, Nagata, Y, Kondoh, O, Ishii, N. | | Deposit date: | 2008-02-22 | | Release date: | 2009-02-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Synthesis and structure-activity relationships of novel benzofuran farnesyltransferase inhibitors
Bioorg.Med.Chem.Lett., 19, 2009
|
|
1RIS
 
 | | CRYSTAL STRUCTURE OF THE RIBOSOMAL PROTEIN S6 FROM THERMUS THERMOPHILUS | | Descriptor: | RIBOSOMAL PROTEIN S6 | | Authors: | Lindahl, M, Svensson, L.A, Liljas, A, Sedelnikova, S.E, Eliseikina, I.A, Fomenkova, N.P, Nevskaya, N, Nikonov, S.V, Garber, M.B, Muranova, T.A, Rykonova, A.I, Amons, R. | | Deposit date: | 1994-05-31 | | Release date: | 1994-09-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the ribosomal protein S6 from Thermus thermophilus.
EMBO J., 13, 1994
|
|
