2M09
 
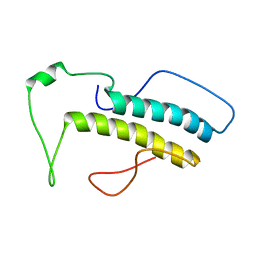 | | Structure, phosphorylation and U2AF65 binding of the Nterminal Domain of splicing factor 1 during 3 splice site Recognition | | Descriptor: | Splicing factor 1 | | Authors: | Madl, T, Sattler, M, Zhang, Y, Bagdiul, I, Kern, T, Kang, H, Zou, P, Maeusbacher, N, Sieber, S.A, Kraemer, A. | | Deposit date: | 2012-10-22 | | Release date: | 2013-01-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, phosphorylation and U2AF65 binding of the N-terminal domain of splicing factor 1 during 3'-splice site recognition.
Nucleic Acids Res., 41, 2013
|
|
8I5B
 
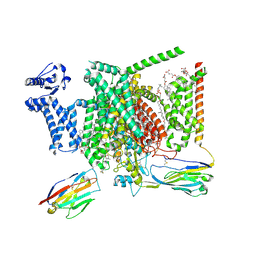 | | Structure of human Nav1.7 in complex with bupivacaine | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-24 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5X
 
 | | Structure of human Nav1.7 in complex with Vinpocetine | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8I5Y
 
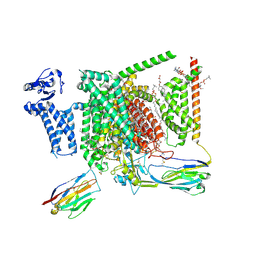 | | Structure of human Nav1.7 in complex with vixotrigine | | Descriptor: | (2S,3R,4E)-2-(acetylamino)-3-hydroxyoctadec-4-en-1-yl dihydrogen phosphate, (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8IGU
 
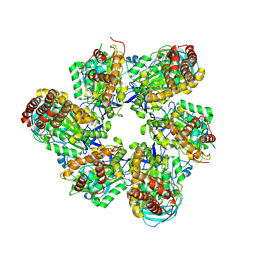 | |
8I5G
 
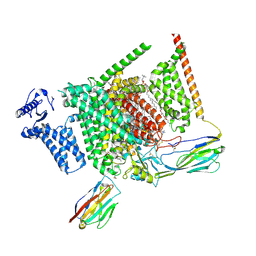 | | Structure of human Nav1.7 in complex with PF-05089771 | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-O-OCTADECYL-SN-GLYCERO-3-PHOSPHOCHOLINE, ... | | Authors: | Wu, Q.R, Yan, N. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structural mapping of Na v 1.7 antagonists.
Nat Commun, 14, 2023
|
|
8IGW
 
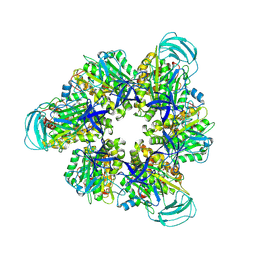 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 4 ADPs: A3(De)3_(ADP)3cat,1non-cat, Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP)3cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, V-type sodium ATPase catalytic subunit A, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
3ZNJ
 
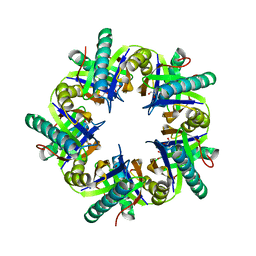 | | Crystal structure of unliganded ClcF from R.opacus 1CP in crystal form 1. | | Descriptor: | 1,2-ETHANEDIOL, 5-CHLOROMUCONOLACTONE DEHALOGENASE, CHLORIDE ION | | Authors: | Roth, C, Groening, J.A.D, Kaschabek, S.R, Schloemann, M, Straeter, N. | | Deposit date: | 2013-02-14 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure and Catalytic Mechanism of Chloromuconolactone Dehalogenase Clcf from Rhodococcus Opacus 1Cp.
Mol.Microbiol., 88, 2013
|
|
8IGV
 
 | | Hexameric Ring Complex of Engineered V1-ATPase bound to 5 ADPs: A3(De)3_(ADP-Pi)1cat(ADP)2cat,2non-cat | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kosugi, T, Tanabe, M, Koga, N. | | Deposit date: | 2023-02-21 | | Release date: | 2023-07-12 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Design of allosteric sites into rotary motor V 1 -ATPase by restoring lost function of pseudo-active sites.
Nat.Chem., 15, 2023
|
|
8IC0
 
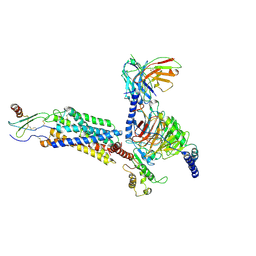 | | Cryo-EM structure of CXCL8 bound C-X-C chemokine receptor 1 in complex with Gi heterotrimer | | Descriptor: | C-X-C chemokine receptor type 1, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Ishimoto, N, Park, J.H, Park, S.Y. | | Deposit date: | 2023-02-10 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of CXC chemokine receptor 1 ligand binding and activation.
Nat Commun, 14, 2023
|
|
6SHQ
 
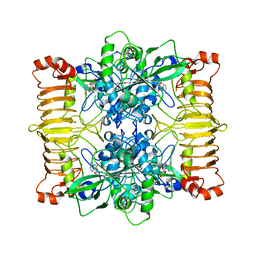 | | Escherichia coli AGPase in complex with AMP. Symmetry C2 | | Descriptor: | ADENOSINE MONOPHOSPHATE, Glucose-1-phosphate adenylyltransferase | | Authors: | Cifuente, J.O, Comino, N, D'Angelo, C, Marina, A, Gil-Carton, D, Albesa-Jove, D, Guerin, M.E. | | Deposit date: | 2019-08-07 | | Release date: | 2020-02-05 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The allosteric control mechanism of bacterial glycogen biosynthesis disclosed by cryoEM.
Curr Res Struct Biol, 2, 2020
|
|
8I7E
 
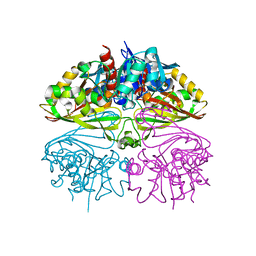 | | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A | | Descriptor: | Glyceraldehyde-3-phosphate dehydrogenase | | Authors: | Kumar, N, Dilawari, R, Chaubey, G.K, Modanwal, R, Talukdar, S, Dhiman, A, Chaudhary, S, Patidar, A, Kumar, A, Raje, C.I, Raje, M, Kumaran, S. | | Deposit date: | 2023-01-31 | | Release date: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of Glyceraldehyde 3-phosphate dehydrogenase from Salmonella typhi at 2.05A
To Be Published
|
|
8ILR
 
 | | Cryo-EM structure of PI3Kalpha in complex with compound 16 | | Descriptor: | N-[(2S)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILV
 
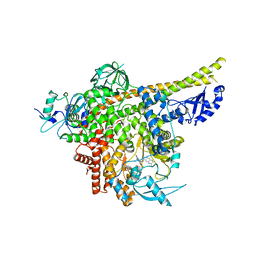 | | Cryo-EM structure of PI3Kalpha in complex with compound 18 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[3-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.19 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8I7P
 
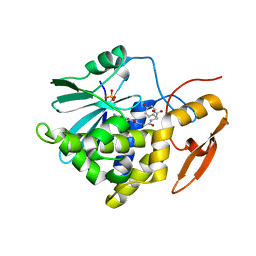 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-tyrosine | | Descriptor: | 6-(2-ethyl-4-hydroxyphenyl)-1H-indazole-3-carboxamide, Ricin A chain, SULFATE ION | | Authors: | Goto, M, Sakamoto, N, Higashi, S, Kawata, R, Nagatsu, K, Saito, R. | | Deposit date: | 2023-02-01 | | Release date: | 2023-09-20 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of ricin toxin A chain complexed with a highly potent pterin-based small-molecular inhibitor.
J Enzyme Inhib Med Chem, 38, 2023
|
|
8ILS
 
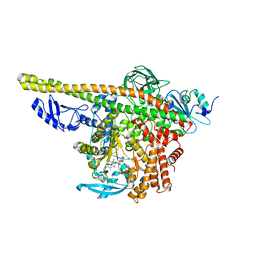 | | Cryo-EM structure of PI3Kalpha in complex with compound 17 | | Descriptor: | N-[(2R)-1-(ethylamino)-1-oxidanylidene-3-[4-(2-quinoxalin-6-ylethynyl)phenyl]propan-2-yl]-2,3-dimethyl-quinoxaline-6-carboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Zhou, Q, Liu, X, Neri, D, Li, W, Favalli, N, Bassi, G, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2023-03-04 | | Release date: | 2023-09-13 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural insights into the interaction of three Y-shaped ligands with PI3K alpha.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ILB
 
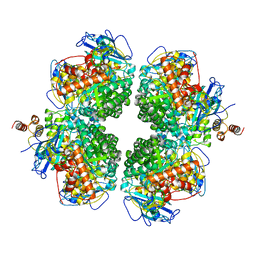 | | The complexes of RbcL, AtRaf1 and AtBSD2 (LFB) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8I4J
 
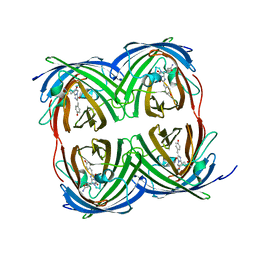 | |
8ILM
 
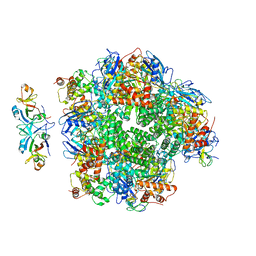 | | The cryo-EM structure of eight Rubisco large subunits (RbcL), two Arabidopsis thaliana Rubisco accumulation factors 1 (AtRaf1), and seven Arabidopsis thaliana Bundle Sheath Defective 2 (AtBSD2) | | Descriptor: | Protein BUNDLE SHEATH DEFECTIVE 2, chloroplastic, Ribulose bisphosphate carboxylase large chain, ... | | Authors: | Wang, R, Song, H, Zhang, W, Wang, N, Zhang, S, Shao, R. | | Deposit date: | 2023-03-03 | | Release date: | 2023-11-01 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the functions of Raf1 and Bsd2 in hexadecameric Rubisco assembly.
Mol Plant, 16, 2023
|
|
8I4K
 
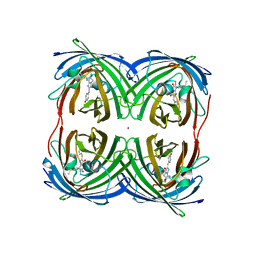 | | Structure of Azami Red1.0, a red fluorescent protein engineered from Azami Green | | Descriptor: | Azami Red1.0, CALCIUM ION | | Authors: | Otsubo, S, Takekawa, N, Imamura, H, Imada, K. | | Deposit date: | 2023-01-19 | | Release date: | 2023-11-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Red fluorescent proteins engineered from green fluorescent proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8HRU
 
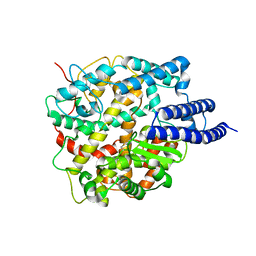 | |
8HRI
 
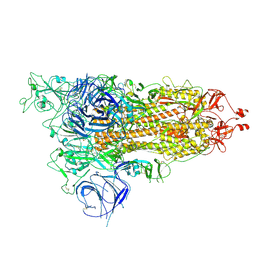 | |
8HRM
 
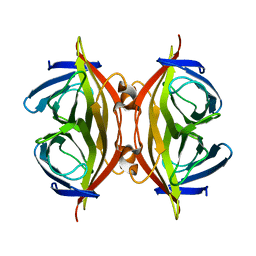 | | Cryo-EM structure of streptavidin | | Descriptor: | Streptavidin | | Authors: | Xu, J, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (2.56 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRK
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 complex | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
8HRL
 
 | | SARS-CoV-2 Delta S-RBD-ACE2 | | Descriptor: | Processed angiotensin-converting enzyme 2, Spike protein S1 | | Authors: | Xu, J, Meng, F, Liu, N, Wang, H.W. | | Deposit date: | 2022-12-15 | | Release date: | 2023-12-20 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Self-assembled monolayers guided free-standing atomic-crystal/molecule superstructure
To Be Published
|
|
