5Y6U
 
 | |
5Y80
 
 | | Complex structure of cyclin G-associated kinase with gefitinib | | Descriptor: | Cyclin-G-associated kinase, Gefitinib, NANOBODY | | Authors: | Ohbayashi, N, Murayama, K, Kato-Murayama, M, Shirouzu, M. | | Deposit date: | 2017-08-18 | | Release date: | 2018-08-29 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis for the Inhibition of Cyclin G-Associated Kinase by Gefitinib.
ChemistryOpen, 7, 2018
|
|
1IF0
 
 | | PSEUDO-ATOMIC MODEL OF BACTERIOPHAGE HK97 PROCAPSID (PROHEAD II) | | Descriptor: | PROTEIN (MAJOR CAPSID PROTEIN GP5) | | Authors: | Conway, J.F, Wikoff, W.R, Cheng, N, Duda, R.L, Hendrix, R.W, Johnson, J.E, Steven, A.C. | | Deposit date: | 2001-04-11 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Virus maturation involving large subunit rotations and local refolding.
Science, 292, 2001
|
|
1F2K
 
 | | CRYSTAL STRUCTURE OF ACANTHAMOEBA CASTELLANII PROFILIN II, CUBIC CRYSTAL FORM | | Descriptor: | PROFILIN II | | Authors: | Fedorov, A.A, Shi, W, Mahoney, N, Kaiser, D.A, Almo, S.C. | | Deposit date: | 2000-05-26 | | Release date: | 2000-06-08 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Comparative Structural Analysis of Profilins
To be Published
|
|
5YDZ
 
 | |
1ITW
 
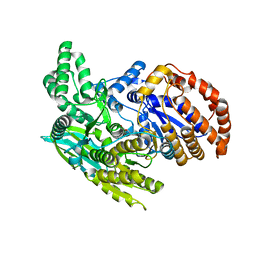 | | Crystal structure of the monomeric isocitrate dehydrogenase in complex with isocitrate and Mn | | Descriptor: | ISOCITRIC ACID, Isocitrate dehydrogenase, MANGANESE (II) ION | | Authors: | Yasutake, Y, Watanabe, S, Yao, M, Takada, Y, Fukunaga, N, Tanaka, I. | | Deposit date: | 2002-02-12 | | Release date: | 2002-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure of the Monomeric Isocitrate Dehydrogenase: Evidence of a Protein Monomerization by a Domain Duplication
Structure, 10, 2002
|
|
6JLR
 
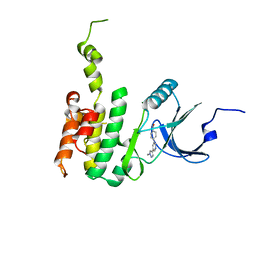 | |
1WZA
 
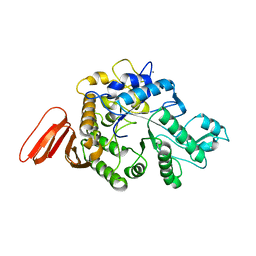 | |
7EET
 
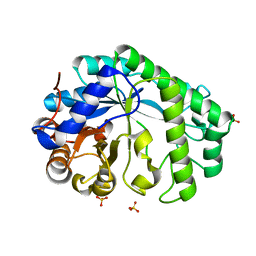 | | Mannanase KMAN from Klebsiella oxytoca KUB-CW2-3 | | Descriptor: | Mannanase KMAN, SULFATE ION | | Authors: | Pongsapipatana, N, Charoenwattanasatien, R, Pramanpol, N, Nitisinprasert, S, Keawsompong, S. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Crystallization, structural characterization and kinetic analysis of a GH26 beta-mannanase from Klebsiella oxytoca KUB-CW2-3.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1IS7
 
 | | Crystal structure of rat GTPCHI/GFRP stimulatory complex | | Descriptor: | GTP Cyclohydrolase I, GTP Cyclohydrolase I Feedback Regulatory Protein, PHENYLALANINE, ... | | Authors: | Maita, N, Okada, K, Hatakeyama, K, Hakoshima, T. | | Deposit date: | 2001-11-18 | | Release date: | 2002-02-20 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the stimulatory complex of GTP cyclohydrolase I and its feedback regulatory protein GFRP.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IKC
 
 | | NMR Structure of alpha-Bungarotoxin | | Descriptor: | long neurotoxin 1 | | Authors: | Niccolai, N, Spiga, O, Ciutti, A. | | Deposit date: | 2001-05-03 | | Release date: | 2001-05-16 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR structure of alpha-bungarotoxin free and bound to a mimotope of the nicotinic acetylcholine receptor.
Biochemistry, 41, 2002
|
|
1X66
 
 | | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor | | Descriptor: | Friend leukemia integration 1 transcription factor | | Authors: | Goroncy, A, Kigawa, T, Koshiba, S, Sato, M, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-17 | | Release date: | 2005-11-17 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the SAM_PNT-domain of the human friend LEUKEMIAINTEGRATION 1 transcription factor
To be Published
|
|
7L0N
 
 | | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity | | Descriptor: | 1-METHOXY-2-[2-(2-METHOXY-ETHOXY]-ETHANE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Snell, G, Czudnochowski, N, Dillen, J, Nix, J.C, Croll, T.I, Corti, D. | | Deposit date: | 2020-12-11 | | Release date: | 2021-02-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Circulating SARS-CoV-2 spike N439K variants maintain fitness while evading antibody-mediated immunity.
Cell, 184, 2021
|
|
1IUB
 
 | | Fucose-specific lectin from Aleuria aurantia (Hg-derivative form) | | Descriptor: | CHLORIDE ION, Fucose-specific lectin, MERCURY (II) ION, ... | | Authors: | Fujihashi, M, Peapus, D.H, Kamiya, N, Nagata, Y, Miki, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-03-01 | | Release date: | 2003-09-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Fucose-Specific Lectin from Aleuria aurantia Binding Ligands at Three of Its Five Sugar Recognition Sites
Biochemistry, 42, 2003
|
|
7NMB
 
 | |
6JTB
 
 | | Crystal structure of dipeptidyl peptidase 11 (DPP11) with citrate from Porphyromonas gingivalis (Space) | | Descriptor: | Asp/Glu-specific dipeptidyl-peptidase, CITRIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sakamoto, Y, Suzuki, Y, Iizuka, I, Roppongi, S, Kushibiki, C, Nakamura, A, Ogasawara, W, Tanaka, N. | | Deposit date: | 2019-04-10 | | Release date: | 2019-10-02 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Fragment-based discovery of the first nonpeptidyl inhibitor of an S46 family peptidase.
Sci Rep, 9, 2019
|
|
1T0W
 
 | | 25 NMR structures of Truncated Hevein of 32 aa (Hevein-32) complex with N,N,N-triacetylglucosamina | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hevein | | Authors: | Aboitiz, N, Vila-Perello, M, Groves, P, Asensio, J.L, Andreu, D, Canada, F.J, Jimenez-Barbero, J. | | Deposit date: | 2004-04-13 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | NMR and modeling studies of protein-carbohydrate interactions: synthesis, three-dimensional structure, and recognition properties of a minimum hevein domain with binding affinity for chitooligosaccharides
Chembiochem, 5, 2004
|
|
6HAY
 
 | | Crystal structure of PROTAC 1 in complex with the bromodomain of human SMARCA2 and pVHL:ElonginC:ElonginB | | Descriptor: | (2~{S},4~{R})-~{N}-[[2-[2-[2-[2-[4-[3-azanyl-6-(2-hydroxyphenyl)pyridazin-4-yl]piperazin-1-yl]ethoxy]ethoxy]ethoxy]-4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-1-[(2~{S})-2-[(1-fluoranylcyclopropyl)carbonylamino]-3,3-dimethyl-butanoyl]-4-oxidanyl-pyrrolidine-2-carboxamide, 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Roy, M, Bader, G, Diers, E, Trainor, N, Farnaby, W, Ciulli, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-06-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | BAF complex vulnerabilities in cancer demonstrated via structure-based PROTAC design.
Nat.Chem.Biol., 15, 2019
|
|
1UBG
 
 | | MsREcA-dATP complex | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, RecA | | Authors: | Datta, S, Krishna, R, Ganesh, N, Chandra, N.R, Muniyappa, K, Vijayan, M. | | Deposit date: | 2003-04-04 | | Release date: | 2003-07-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal Structures of Mycobacterium smegmatis RecA and Its Nucleotide Complexes
J.BACTERIOL., 185, 2003
|
|
6HDY
 
 | | Crystal structure of 2-Hydroxyisobutyryl-CoA Ligase (HCL) in the postadenylation state in complex with S3-HB-AMP | | Descriptor: | (3S)-3-HYDROXYBUTANOIC ACID, 2-hydroxyisobutyryl-CoA synthetase, SULFATE ION, ... | | Authors: | Zahn, M, Rohwerder, T, Strater, N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of 2-Hydroxyisobutyric Acid-CoA Ligase Reveal Determinants of Substrate Specificity and Describe a Multi-Conformational Catalytic Cycle.
J.Mol.Biol., 431, 2019
|
|
1UDM
 
 | | Solution structure of Coactosin-like protein (Cofilin family) from Mus Musculus | | Descriptor: | Coactosin-like protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-01 | | Release date: | 2004-05-18 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | NMR solution structures of actin depolymerizing factor homology domains.
Protein Sci., 18, 2009
|
|
1JB6
 
 | |
1JEI
 
 | | LEM DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN EMERIN | | Descriptor: | EMERIN | | Authors: | Wolff, N, Gilquin, B, Courchay, K, Callebaut, I, Zinn-Justin, S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of emerin, an inner nuclear membrane protein mutated in X-linked Emery-Dreifuss muscular dystrophy
FEBS LETT., 501, 2001
|
|
5ZC2
 
 | | Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH), reductase component (C1) | | Descriptor: | FLAVIN MONONUCLEOTIDE, p-hydroxyphenylacetate 3-hydroxylase, reductase component | | Authors: | Yuenyao, A, Petchyam, N, Chaiyen, P, Pakotiprapha, D. | | Deposit date: | 2018-02-14 | | Release date: | 2018-08-08 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.898 Å) | | Cite: | Crystal structure of the flavin reductase of Acinetobacter baumannii p-hydroxyphenylacetate 3-hydroxylase (HPAH) and identification of amino acid residues underlying its regulation by aromatic ligands
Arch. Biochem. Biophys., 653, 2018
|
|
1K30
 
 | | Crystal Structure Analysis of Squash (Cucurbita moschata) glycerol-3-phosphate (1)-acyltransferase | | Descriptor: | glycerol-3-phosphate acyltransferase | | Authors: | Turnbull, A.P, Rafferty, J.B, Sedelnikova, S.E, Slabas, A.R, Schierer, T.P, Kroon, J.T, Simon, J.W, Fawcett, T, Nishida, I, Murata, N, Rice, D.W. | | Deposit date: | 2001-10-01 | | Release date: | 2001-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Analysis of the structure, substrate specificity, and mechanism of squash glycerol-3-phosphate (1)-acyltransferase.
Structure, 9, 2001
|
|
