4PID
 
 | | Crystal structure of human adenovirus 2 protease with a weak pyrimidine nitrile inhibitor | | 分子名称: | ACETATE ION, N-benzyl-2-[(Z)-iminomethyl]pyrimidine-5-carboxamide, Pre-protein VI, ... | | 著者 | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | 登録日 | 2014-05-08 | | 公開日 | 2014-09-10 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.59 Å) | | 主引用文献 | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
6BUU
 
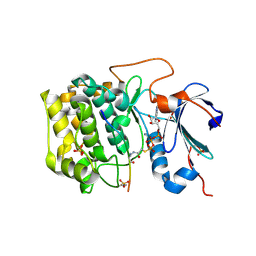 | | Crystal structure of AKT1 (aa 144-480) with a bisubstrate | | 分子名称: | GLY-ARG-PRO-ARG-THR-THR-ZXW-PHE-ALA-GLU, MANGANESE (II) ION, RAC-alpha serine/threonine-protein kinase, ... | | 著者 | Chu, N, Gabelli, S.B, Cole, P.A. | | 登録日 | 2017-12-11 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-07-10 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Akt Kinase Activation Mechanisms Revealed Using Protein Semisynthesis.
Cell, 174, 2018
|
|
4PIQ
 
 | | Crystal structure of human adenovirus 8 protease with a nitrile inhibitor | | 分子名称: | N-[(3,5-dichlorophenyl)acetyl]-L-threonyl-N-[(2Z)-2-iminoethyl]glycinamide, PVI, Protease | | 著者 | Mac Sweeney, A, Grosche, P, Ellis, D, Combrink, K, Erbel, P, Hughes, N, Sirockin, F, Melkko, S, Bernardi, A, Ramage, P, Jarousse, N, Altmann, E. | | 登録日 | 2014-05-09 | | 公開日 | 2014-09-10 | | 最終更新日 | 2023-12-27 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Discovery and structure-based optimization of adenain inhibitors.
Acs Med.Chem.Lett., 5, 2014
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | 登録日 | 2023-12-04 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.7 Å) | | 主引用文献 | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
6C16
 
 | |
6C1M
 
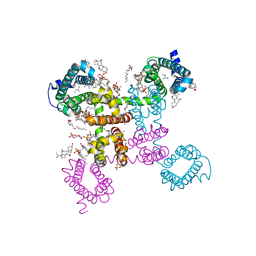 | | NavAb NormoPP mutant | | 分子名称: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1-METHYLGUANIDINE, CHAPSO, ... | | 著者 | Catterall, W.A, Zheng, N, Jiang, D, Gamal El-Din, T.M. | | 登録日 | 2018-01-04 | | 公開日 | 2018-05-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.518 Å) | | 主引用文献 | Structural basis for gating pore current in periodic paralysis.
Nature, 557, 2018
|
|
6RV2
 
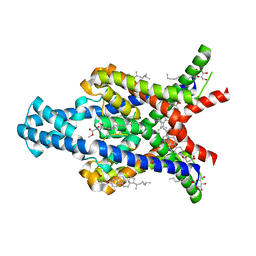 | | Crystal structure of the human two pore domain potassium ion channel TASK-1 (K2P3.1) in a closed conformation | | 分子名称: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DECYL-BETA-D-MALTOPYRANOSIDE, ... | | 著者 | Rodstrom, K.E.J, Pike, A.C.W, Zhang, W, Quigley, A, Speedman, D, Mukhopadhyay, S.M.M, Shrestha, L, Chalk, R, Venkaya, S, Bushell, S.R, Tessitore, A, Burgess-Brown, N, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Carpenter, E.P, Structural Genomics Consortium (SGC) | | 登録日 | 2019-05-30 | | 公開日 | 2019-08-07 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | A lower X-gate in TASK channels traps inhibitors within the vestibule.
Nature, 582, 2020
|
|
8UUB
 
 | | Structure of hypothiocyanous acid reductase (Har) from Streptococcus pneumoniae | | 分子名称: | FLAVIN-ADENINE DINUCLEOTIDE, MALONATE ION, Oxidoreductase, ... | | 著者 | Shearer, H.L, Currie, M.J, Dickerhof, N, Dobson, R.C.J. | | 登録日 | 2023-10-31 | | 公開日 | 2024-04-17 | | 最終更新日 | 2024-05-22 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Hypothiocyanous acid reductase is critical for host colonization and infection by Streptococcus pneumoniae.
J.Biol.Chem., 300, 2024
|
|
6BJG
 
 | | CIRV p19 mutant T111H in complex with siRNA | | 分子名称: | RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3'), RNA silencing suppressor p19 | | 著者 | Foss, D.V, Schirle, N, Pezacki, J.P, Macrae, I.J. | | 登録日 | 2017-11-06 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs.
Febs Open Bio, 9, 2019
|
|
8V85
 
 | |
6S6B
 
 | | Type III-B Cmr-beta Cryo-EM structure of the Apo state | | 分子名称: | CRISPR-associated RAMP protein, Cmr4 family, Cmr6 family, ... | | 著者 | Sofos, N, Montoya, G, Stella, S. | | 登録日 | 2019-07-02 | | 公開日 | 2020-07-08 | | 最終更新日 | 2020-09-16 | | 実験手法 | ELECTRON MICROSCOPY (2.75 Å) | | 主引用文献 | Structures of the Cmr-beta Complex Reveal the Regulation of the Immunity Mechanism of Type III-B CRISPR-Cas.
Mol.Cell, 79, 2020
|
|
8V83
 
 | | 60S ribosome biogenesis intermediate (Dbp10 pre-catalytic structure - Overall map) | | 分子名称: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | 著者 | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | 登録日 | 2023-12-04 | | 公開日 | 2024-05-01 | | 実験手法 | ELECTRON MICROSCOPY (2.53 Å) | | 主引用文献 | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
7Z19
 
 | | E. coli C-P lyase bound to a single PhnK ABC domain | | 分子名称: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | 著者 | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | 登録日 | 2022-02-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z18
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer and ATP | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, ... | | 著者 | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | 登録日 | 2022-02-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (1.98 Å) | | 主引用文献 | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
6BJV
 
 | | CIRV p19 protein in complex with siRNA | | 分子名称: | RNA (5'-R(P*CP*GP*UP*AP*CP*GP*CP*GP*GP*AP*AP*UP*AP*CP*UP*UP*CP*GP*AP*UP*U)-3'), RNA (5'-R(P*UP*CP*GP*AP*AP*GP*UP*AP*UP*UP*CP*CP*GP*CP*GP*UP*AP*CP*GP*UP*U)-3'), RNA silencing suppressor p19 | | 著者 | Foss, D.V, Schirle, N, Pezacki, J.P, Macrae, I.J. | | 登録日 | 2017-11-07 | | 公開日 | 2019-01-16 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.198 Å) | | 主引用文献 | Structural insights into interactions between viral suppressor of RNA silencing protein p19 mutants and small RNAs.
Febs Open Bio, 9, 2019
|
|
7Z17
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | 分子名称: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | 著者 | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | 登録日 | 2022-02-24 | | 公開日 | 2022-05-25 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (2.57 Å) | | 主引用文献 | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z15
 
 | | E. coli C-P lyase bound to a PhnK/PhnL dual ABC dimer and ADP + Pi | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, ... | | 著者 | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | 登録日 | 2022-02-24 | | 公開日 | 2022-06-22 | | 最終更新日 | 2024-07-17 | | 実験手法 | ELECTRON MICROSCOPY (1.93 Å) | | 主引用文献 | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
8V4K
 
 | | CCP5 in complex with microtubules class1 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.1 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3N
 
 | | CCP5 in complex with Glu-P-Glu transition state analog | | 分子名称: | (2S)-2-{[(S)-[(3S)-3-acetamido-4-(ethylamino)-4-oxobutyl](hydroxy)phosphoryl]methyl}pentanedioic acid, Cytosolic carboxypeptidase-like protein 5, D-MALATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3O
 
 | | CCP5 in complex with Glu-P-peptide 1 transition state analog | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, POTASSIUM ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V4M
 
 | | CCP5 in complex with microtubules class3 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-29 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-10-23 | | 実験手法 | ELECTRON MICROSCOPY (3 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3S
 
 | | Structure of CCP5 class3 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3M
 
 | | CCP5 apo structure | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, D-MALATE, IMIDAZOLE, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
8V3R
 
 | | Structure of CCP5 class2 | | 分子名称: | Cytosolic carboxypeptidase-like protein 5, GLUTAMIC ACID, ZINC ION, ... | | 著者 | Chen, J, Zehr, E.A, Gruschus, J.M, Szyk, A, Liu, Y, Tanner, M.E, Tjandra, N, Roll-Mecak, A. | | 登録日 | 2023-11-28 | | 公開日 | 2024-07-17 | | 最終更新日 | 2024-08-07 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Tubulin code eraser CCP5 binds branch glutamates by substrate deformation.
Nature, 631, 2024
|
|
7JV4
 
 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (one RBD open) | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | 著者 | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | 登録日 | 2020-08-20 | | 公開日 | 2020-10-14 | | 最終更新日 | 2024-10-16 | | 実験手法 | ELECTRON MICROSCOPY (3.4 Å) | | 主引用文献 | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
