6PBC
 
 | | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma,1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1, CALCIUM ION, SODIUM ION | | Authors: | Hajicek, N, Sondek, J. | | Deposit date: | 2019-06-13 | | Release date: | 2020-01-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structural basis for the activation of PLC-gamma isozymes by phosphorylation and cancer-associated mutations.
Elife, 8, 2019
|
|
5GYY
 
 | | Plant receptor complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, S-locus protein 11, S-receptor kinase SRK9 | | Authors: | Ma, R, Han, Z, Hu, Z, Lin, G, Gong, X, Zhang, H, June, N, Chai, J. | | Deposit date: | 2016-09-24 | | Release date: | 2017-09-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.351 Å) | | Cite: | Plant receptor complex at 2.35 Angstroms resolution
To Be Published
|
|
6P6Y
 
 | | Crystal structure of voltage-gated sodium channel NavAb V100C/Q150C disulfide crosslinked mutant in the activated state | | Descriptor: | 1,2-DIMYRISTOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, Ion transport protein | | Authors: | Wisedchaisri, G, Tonggu, L, McCord, E, Gamal El-din, T.M, Wang, L, Zheng, N, Catterall, W.A. | | Deposit date: | 2019-06-04 | | Release date: | 2019-08-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Resting-State Structure and Gating Mechanism of a Voltage-Gated Sodium Channel.
Cell, 178, 2019
|
|
1J05
 
 | | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment | | Descriptor: | GLYCEROL, PHOSPHATE ION, anti-CEA mAb T84.66, ... | | Authors: | Kondo, H, Nishimura, Y, Shiroishi, M, Asano, R, Noro, N, Tsumoto, K, Kumagai, I. | | Deposit date: | 2002-11-01 | | Release date: | 2003-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of anti-carcinoembryonic antigen monoclonal antibody T84.66 Fv fragment
To be Published
|
|
5H0R
 
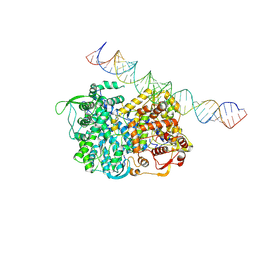 | | RNA dependent RNA polymerase ,vp4,dsRNA | | Descriptor: | RNA (42-MER), RNA-dependent RNA polymerase, VP4 protein | | Authors: | Li, X, Zhou, N, Chen, W, Zhu, B, Wang, X, Xu, B, Wang, J, Liu, H, Cheng, L. | | Deposit date: | 2016-10-06 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Near-Atomic Resolution Structure Determination of a Cypovirus Capsid and Polymerase Complex Using Cryo-EM at 200kV
J. Mol. Biol., 429, 2017
|
|
6LF5
 
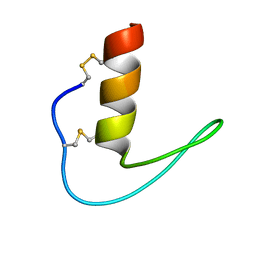 | | The solution structure of ShSPI | | Descriptor: | ShSPI | | Authors: | Luan, N, Rong, M.Q, Liu, J.X, Lai, R. | | Deposit date: | 2019-11-29 | | Release date: | 2020-12-02 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Identification and Characterization of ShSPI, a Kazal-Type Elastase Inhibitor from the Venom of Scolopendra Hainanum .
Toxins, 11, 2019
|
|
1IWL
 
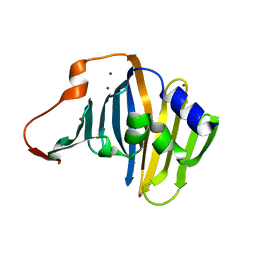 | | Crystal Structure of the Lipoprotein Localization Factor, LolA | | Descriptor: | MAGNESIUM ION, Outer-membrane lipoproteins carrier protein, ZINC ION | | Authors: | Takeda, K, Miyatake, H, Yokota, N, Matsuyama, S, Tokuda, H, Miki, K. | | Deposit date: | 2002-05-17 | | Release date: | 2003-07-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of bacterial lipoprotein localization factors, LolA and LolB
Embo J., 22, 2003
|
|
5GKY
 
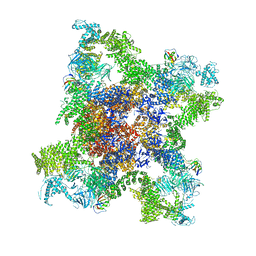 | | Structure of RyR1 in a closed state (C1 conformer) | | Descriptor: | Peptidyl-prolyl cis-trans isomerase FKBP1A, Ryanodine receptor 1, ZINC ION | | Authors: | Bai, X.C, Yan, Z, Wu, J.P, Yan, N. | | Deposit date: | 2016-07-07 | | Release date: | 2016-08-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | The Central domain of RyR1 is the transducer for long-range allosteric gating of channel opening
Cell Res., 26, 2016
|
|
1IWU
 
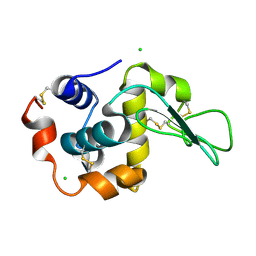 | | Crystal Structure Analysis of Human lysozyme at 127K. | | Descriptor: | CHLORIDE ION, LYSOZYME C | | Authors: | Joti, Y, Nakasako, M, Kidera, A, Go, N. | | Deposit date: | 2002-06-03 | | Release date: | 2002-09-04 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Nonlinear temperature dependence of the crystal structure of lysozyme: correlation between coordinate shifts and thermal factors.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
6PPR
 
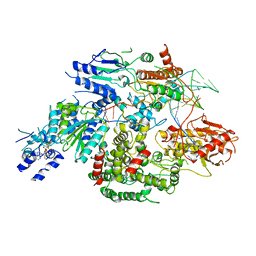 | | Cryo-EM structure of AdnA(D934A)-AdnB(D1014A) in complex with AMPPNP and DNA | | Descriptor: | ATP-dependent DNA helicase (UvrD/REP), DNA (70-MER), IRON/SULFUR CLUSTER, ... | | Authors: | Jia, N, Unciuleac, M, Shuman, S, Patel, D.J. | | Deposit date: | 2019-07-08 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structures and single-molecule analysis of bacterial motor nuclease AdnAB illuminate the mechanism of DNA double-strand break resection.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6KVG
 
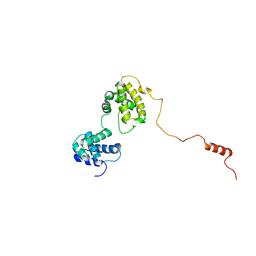 | | The solution structure of human Orc6 | | Descriptor: | Origin recognition complex subunit 6 | | Authors: | Liu, C, Xu, N, You, Y, Zhu, G. | | Deposit date: | 2019-09-04 | | Release date: | 2020-09-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of DNA replication origin recognition by human Orc6 protein binding with DNA.
Nucleic Acids Res., 48, 2020
|
|
1IYF
 
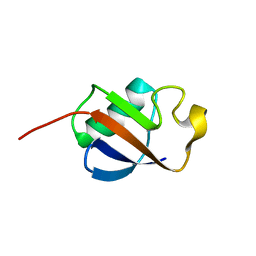 | | Solution structure of ubiquitin-like domain of human parkin | | Descriptor: | parkin | | Authors: | Sakata, E, Yamaguchi, Y, Kurimoto, E, Kikuchi, J, Yokoyama, S, Kawahara, H, Yokosawa, H, Hattori, N, Mizuno, Y, Tanaka, K, Kato, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2002-08-13 | | Release date: | 2003-03-25 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Parkin binds the Rpn10 subunit of 26S proteasomes through its ubiquitin-like domain
EMBO REP., 4, 2003
|
|
1J3Z
 
 | | Direct observation of photolysis-induced tertiary structural changes in human haemoglobin; Crystal structure of alpha(Fe-CO)-beta(Ni) hemoglobin (laser unphotolysed) | | Descriptor: | BUT-2-ENEDIAL, CARBON MONOXIDE, Hemoglobin alpha Chain, ... | | Authors: | Adachi, S, Park, S.-Y, Tame, J.R.H, Shiro, Y, Shibayama, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-21 | | Release date: | 2003-07-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Direct observation of photolysis-induced tertiary structural changes in hemoglobin
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1WFQ
 
 | | Solution structure of the first cold-shock domain of the human KIAA0885 protein (UNR protein) | | Descriptor: | UNR protein | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Tomizawa, T, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-05-26 | | Release date: | 2004-11-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | The NMR solution structures of the five constituent cold-shock domains (CSD) of the human UNR (upstream of N-ras) protein.
J.Struct.Funct.Genom., 11, 2010
|
|
5GIZ
 
 | | Periplasmic heme-binding protein BhuT in apo form | | Descriptor: | CHLORIDE ION, Putative hemin transport system, substrate-binding protein, ... | | Authors: | Nakamura, N, Naoe, Y, Rahman, M.M, Shiro, Y, Sugimoto, H. | | Deposit date: | 2016-06-26 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for binding and transfer of heme in bacterial heme-acquisition systems.
Proteins, 85, 2017
|
|
7Z18
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer and ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
7Z19
 
 | | E. coli C-P lyase bound to a single PhnK ABC domain | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
1J7H
 
 | | Solution Structure of HI0719, a Hypothetical Protein From Haemophilus Influenzae | | Descriptor: | HYPOTHETICAL PROTEIN HI0719 | | Authors: | Parsons, L, Bonander, N, Eisenstein, E, Gilson, M, Kairys, V, Orban, J, Structure 2 Function Project (S2F) | | Deposit date: | 2001-05-16 | | Release date: | 2003-02-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Functional Ligand Screening of HI0719, a Highly Conserved Protein from Bacteria to Humans in the YjgF/YER057c/UK114 Family
Biochemistry, 42, 2003
|
|
7Z17
 
 | | E. coli C-P lyase bound to a PhnK ABC dimer in an open conformation | | Descriptor: | Alpha-D-ribose 1-methylphosphonate 5-phosphate C-P lyase, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnG, Alpha-D-ribose 1-methylphosphonate 5-triphosphate synthase subunit PhnH, ... | | Authors: | Amstrup, S.K, Sofos, N, Karlsen, J.L, Skjerning, R.B, Boesen, T, Enghild, J.J, Hove-Jensen, B, Brodersen, D.E. | | Deposit date: | 2022-02-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.57 Å) | | Cite: | Structural remodelling of the carbon-phosphorus lyase machinery by a dual ABC ATPase.
Nat Commun, 14, 2023
|
|
5GLM
 
 | | Crystal structure of CoXyl43, GH43 beta-xylosidase/alpha-arabinofuranosidase from a compost microbial metagenome in complex with xylotriose, calcium-free form. | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Glycoside hydrolase family 43, ... | | Authors: | Matsuzawa, T, Kishine, N, Fujimoto, Z, Yaoi, K. | | Deposit date: | 2016-07-12 | | Release date: | 2017-03-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of metagenomic beta-xylosidase/ alpha-l-arabinofuranosidase activated by calcium.
J. Biochem., 162, 2017
|
|
1WWY
 
 | | Solution structure of the DUF1000 domain of a thioredoxin-like protein 1 | | Descriptor: | Thioredoxin-like protein 1 | | Authors: | Goroncy, A.K, Kigawa, T, Koshiba, S, Kobayashi, N, Tochio, N, Inoue, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-01-18 | | Release date: | 2005-07-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the C-terminal DUF1000 domain of the human thioredoxin-like 1 protein.
Proteins, 78, 2010
|
|
6PDN
 
 | | Human PIM1 bound to benzothiophene inhibitor 292 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxylic acid, GLYCEROL, Serine/threonine-protein kinase pim-1, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
6PDO
 
 | | Human PIM1 bound to benzothiophene inhibitor 354 | | Descriptor: | 4-{5-[(3-aminopropyl)carbamoyl]thiophen-2-yl}-1-benzothiophene-2-carboxamide, Peptide, Serine/threonine-protein kinase pim-1 | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Human PIM1
To Be Published
|
|
7ZEM
 
 | |
7ZEO
 
 | |
