6D4M
 
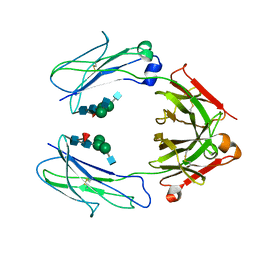 | | Crystal Structure of a Fc Fragment of Rhesus macaque (Macaca mulatta) IgG3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Fc fragment of IgG3 | | Authors: | Gohain, N, Tolbert, W.D, Pazgier, M. | | Deposit date: | 2018-04-18 | | Release date: | 2019-05-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.47 Å) | | Cite: | From Rhesus macaque to human: structural evolutionary pathways for immunoglobulin G subclasses.
Mabs, 11, 2019
|
|
6GBU
 
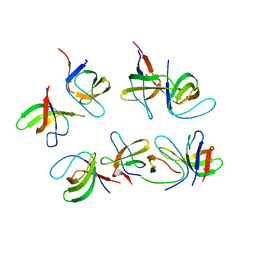 | | Crystal structure of the second SH3 domain of FCHSD2 (SH3-2) in complex with the fourth SH3 domain of ITSN1 (SH3d) | | Descriptor: | F-BAR and double SH3 domains protein 2, Intersectin-1 | | Authors: | Almeida-Souza, L, Frank, R, Garcia-Nafria, J, Colussi, A, Gunawardana, N, Johnson, C.M, Yu, M, Howard, G, Andrews, B, Vallis, Y, McMahon, H.T. | | Deposit date: | 2018-04-16 | | Release date: | 2018-06-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.44 Å) | | Cite: | A Flat BAR Protein Promotes Actin Polymerization at the Base of Clathrin-Coated Pits.
Cell, 174, 2018
|
|
1PE7
 
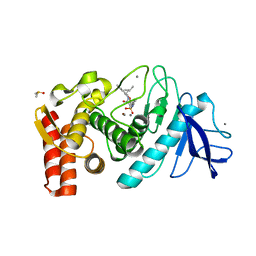 | | Thermolysin with bicyclic inhibitor | | Descriptor: | 2-(4-METHYLPHENOXY)ETHYLPHOSPHINATE, 3-METHYLBUTAN-1-AMINE, CALCIUM ION, ... | | Authors: | Juers, D, Yusuff, N, Bartlett, P.A, Matthews, B.W. | | Deposit date: | 2003-05-21 | | Release date: | 2004-06-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Conformational Constraint and Structural Complementarity in Thermolysin Inhibitors: Structures of Enzyme Complexes and Conclusions
To be Published
|
|
1PHQ
 
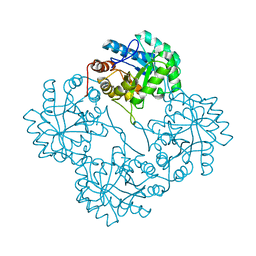 | | Crystal structure of KDO8P synthase in its binary complex with substrate analog E-FPEP | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, 3-FLUORO-2-(PHOSPHONOOXY)PROPANOIC ACID | | Authors: | Vainer, R, Adir, N, Baasov, T, Belakhov, V, Rabkin, E. | | Deposit date: | 2003-05-29 | | Release date: | 2004-07-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystallographic Analysis of the Phosphoenol Pyruvate Binding Site in E. Coli KDO8P Synthase
To be Published
|
|
6FMR
 
 | | IMISX-EP of Se-PepTSt | | Descriptor: | (2S)-2,3-DIHYDROXYPROPYL(7Z)-PENTADEC-7-ENOATE, Di-or tripeptide:H+ symporter, PHOSPHATE ION | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-08-29 | | Last modified: | 2018-10-10 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
5DQ5
 
 | | Endothiapepsin in complex with fragment 209 | | Descriptor: | 3-[2-(4-methyl-1,2-oxazol-3-yl)ethyl]pyridine, ACETATE ION, Endothiapepsin, ... | | Authors: | Radeva, N, Heine, A, Klebe, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
to be published
|
|
1R7G
 
 | | NMR structure of the membrane anchor domain (1-31) of the nonstructural protein 5A (NS5A) of hepatitis C virus (Minimized average structure, Sample in 100mM DPC) | | Descriptor: | Genome polyprotein | | Authors: | Penin, F, Brass, V, Appel, N, Ramboarina, S, Montserret, R, Ficheux, D, Blum, H.E, Bartenschlager, R, Moradpour, D. | | Deposit date: | 2003-10-21 | | Release date: | 2004-08-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure and function of the membrane anchor domain of hepatitis C virus nonstructural protein 5A.
J.Biol.Chem., 279, 2004
|
|
6FMW
 
 | | IMISX-EP of Hg-BacA cocrystallization | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Howe, N, Warshamanage, R, Weinert, T, Panepucci, E, Vogeley, L, Basu, S, Diederichs, K, Caffrey, M, Wang, M. | | Deposit date: | 2018-02-02 | | Release date: | 2018-09-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | In situ serial crystallography for rapid de novo membrane protein structure determination.
Commun Biol, 1, 2018
|
|
6V0G
 
 | | Lipophilic Envelope-spanning Tunnel B (LetB), Model 5 | | Descriptor: | Intermembrane transport protein YebT | | Authors: | Isom, G.L, Coudray, N, MacRae, M.R, McManus, C.T, Ekiert, D.C, Bhabha, G. | | Deposit date: | 2019-11-18 | | Release date: | 2020-05-06 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | LetB Structure Reveals a Tunnel for Lipid Transport across the Bacterial Envelope.
Cell, 181, 2020
|
|
6V9Q
 
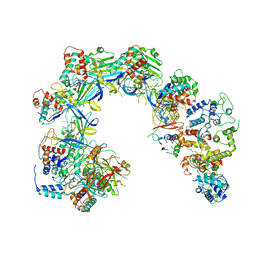 | | Cryo-EM structure of Cascade-TniQ binary complex | | Descriptor: | Cas8, RNA (61-MER), TniQ family protein, ... | | Authors: | Jia, N, Patel, D.J. | | Deposit date: | 2019-12-15 | | Release date: | 2020-01-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure-function insights into the initial step of DNA integration by a CRISPR-Cas-Transposon complex.
Cell Res., 30, 2020
|
|
6VA8
 
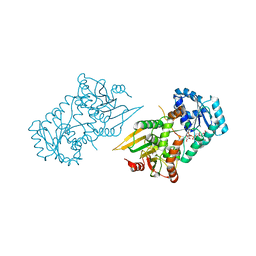 | |
6DEI
 
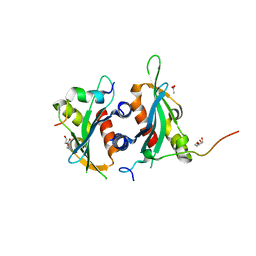 | | Structure of Dse3-Csm1 complex | | Descriptor: | ACETATE ION, Monopolin complex subunit CSM1, Protein DSE3, ... | | Authors: | Singh, N, Corbett, K.D. | | Deposit date: | 2018-05-12 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The budding-yeast RWD protein Csm1 scaffolds diverse protein complexes through a conserved structural mechanism.
Protein Sci., 27, 2018
|
|
6VDI
 
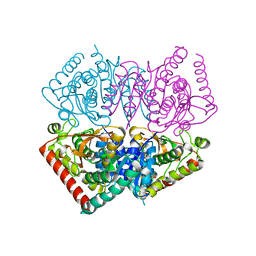 | |
6FUI
 
 | | Complement factor D in complex with the inhibitor 3-((3-((3-(aminomethyl)phenyl)amino)-1H-pyrazolo[3,4-d]pyrimidin-4-yl)amino)phenol | | Descriptor: | (1~{R},2~{S})-2-[[4-[[3-(aminomethyl)phenyl]amino]quinazolin-2-yl]amino]cyclohexane-1-carboxylic acid, Complement factor D | | Authors: | Mac Sweeney, A, Ostermann, N, Vulpetti, A, Maibaum, J, Erbel, P, Lorthiois, E, Yoon, T, Randl, S, Ruedisser, S. | | Deposit date: | 2018-02-27 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Discovery and Design of First Benzylamine-Based Ligands Binding to an Unlocked Conformation of the Complement Factor D.
ACS Med Chem Lett, 9, 2018
|
|
6PNT
 
 | |
6FXU
 
 | |
1RP8
 
 | | Crystal structure of barley alpha-amylase isozyme 1 (amy1) inactive mutant d180a in complex with maltoheptaose | | Descriptor: | Alpha-amylase type 1 isozyme, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2003-12-03 | | Release date: | 2005-06-07 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Oligosaccharide Binding to Barley {alpha}-Amylase 1
J.Biol.Chem., 280, 2005
|
|
8WZ4
 
 | | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 (localized refinement) | | Descriptor: | 5B11 Fab Heavy Chain, 5B11 Fab Light Chain, RSV Fusion glycoprotein | | Authors: | Liu, L, Sun, H, Sun, Y, Zheng, Q, Li, S, Zheng, Z, Xia, N. | | Deposit date: | 2023-11-01 | | Release date: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Cryo-EM structure of prefusion-stabilized RSV F (DS-Cav1 strain: A2) in complex with nAb 5B11 (localized refinement)
To Be Published
|
|
5DPJ
 
 | | sfGFP double mutant - 133/149 p-ethynyl-L-phenylalanine | | Descriptor: | Green fluorescent protein | | Authors: | Dippel, A.B, Olenginski, G.M, Maurici, N, Liskov, M.T, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2015-09-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the effectiveness of spectroscopic reporter unnatural amino acids: a structural study.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5DPZ
 
 | | Endothiapepsin in complex with fragment 31 | | Descriptor: | 2-methylaniline, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Radeva, N, Uehlein, M, Weiss, M, Heine, A, Klebe, G. | | Deposit date: | 2015-09-14 | | Release date: | 2016-09-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.328 Å) | | Cite: | Crystallographic Fragment Screening of an Entire Library
To Be Published
|
|
5DQB
 
 | | Green/cyan WasCFP at pH 8.0 | | Descriptor: | GLYCEROL, SODIUM ION, WasCFP_pH2 | | Authors: | Pletnev, V, Pletneva, N, Pletnev, S. | | Deposit date: | 2015-09-14 | | Release date: | 2016-07-27 | | Last modified: | 2019-02-20 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structure of pH and T dependent green fluorescent protein WasCFP with Trp based chromophore
Russ.J.Bioorganic Chem., 42 (6), 2016
|
|
6UYF
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2mc3-v1 redesigned core from genotype 6a bound to broadly neutralizing antibody AR3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Zhu, J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Proof of concept for rational design of hepatitis C virus E2 core nanoparticle vaccines.
Sci Adv, 6, 2020
|
|
6FZL
 
 | |
8RZF
 
 | |
6PJJ
 
 | | Human PRPF4B bound to benzothiophene inhibitor 224 | | Descriptor: | 1,2-ETHANEDIOL, 4-(5-{[(3-aminophenyl)methyl]carbamoyl}thiophen-2-yl)-1-benzothiophene-2-carboxamide, PHOSPHATE ION, ... | | Authors: | Godoi, P.H.C, Santiago, A.S, Fala, A.M, Ramos, P.Z, Sriranganadane, D, Mascarello, A, Segretti, N, Azevedo, H, Guimaraes, C.R.W, Arruda, P, Elkins, J.M, Counago, R.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2019-06-28 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | to be published
To Be Published
|
|
