4QSD
 
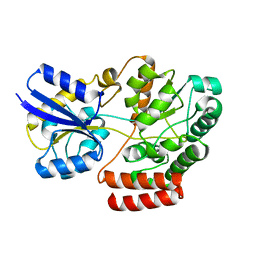 | | Crystal structure of atu4361 sugar transporter from Agrobacterium Fabrum C58, target efi-510558, with bound sucrose | | Descriptor: | ABC-TYPE SUGAR TRANSPORTER, beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-07-03 | | Release date: | 2014-08-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Crystal Structure of Maltoside Transporter Atu4361 from Agrobacterium Fabrum, Target Efi-510558
To be Published
|
|
1OXE
 
 | | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins | | Descriptor: | cyan fluorescent protein cfp | | Authors: | Hyun Bae, J, Rubini, M, Jung, G, Wiegand, G, Seifert, M.H, Azim, M.K, Kim, J.S, Zumbusch, A, Holak, T.A, Moroder, L, Huber, R, Budisa, N. | | Deposit date: | 2003-04-02 | | Release date: | 2003-12-02 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Expansion of the Genetic Code Enables Design of a Novel "Gold" Class of Green Fluorescent Proteins
J.Mol.Biol., 328, 2003
|
|
1P4H
 
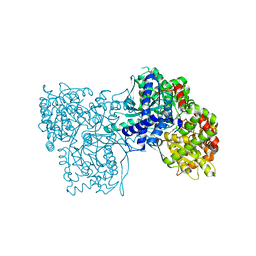 | | Crystal structure of glycogen phosphorylase b in complex with C-(1-acetamido-alpha-D-glucopyranosyl) formamide | | Descriptor: | 1-DEOXY-1-ACETYLAMINO-BETA-D-GLUCO-2-HEPTULOPYRANOSONAMIDE, Glycogen phosphorylase, muscle form, ... | | Authors: | Chrysina, E.D, Oikonomakos, N.G, Zographos, S.E, Kosmopoulou, M.N, Bischler, N, Leonidas, D.D, Nagy, V, Somsak, L, Gergely, P, Praly, J.-P. | | Deposit date: | 2003-04-23 | | Release date: | 2004-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Crsytallographic Studies on alpha- and beta-D-glucopyranosyl Formamide Analogues, Inhibitors of Glycogen Phosphorylase
Biocatal.Biotransfor., 21, 2003
|
|
4R2B
 
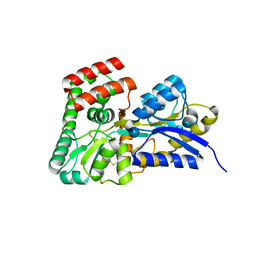 | | Crystal structure of sugar transporter Oant_3817 from Ochrobactrum anthropi, target EFI-510528, with bound glucose | | Descriptor: | Extracellular solute-binding protein family 1, alpha-D-glucopyranose | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Chamala, S, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-11 | | Release date: | 2014-08-27 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal Structure of Glucose Transporter Oant_3817 from Ochrobactrum Anthropi, Target EFI-510528
To be Published
|
|
5OGA
 
 | | Structure of minimal i-motif domain | | Descriptor: | DNA (5'-D(*TP*(DCP)P*GP*TP*TP*CP*(DCP)P*GP*TP*TP*TP*TP*TP*CP*GP*TP*TP*CP*CP*GP*T)-3') | | Authors: | Mir, B, Serrano, I, Buitrago, D, Orozco, M, Escaja, N, Gonzalez, C. | | Deposit date: | 2017-07-12 | | Release date: | 2017-11-29 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Prevalent Sequences in the Human Genome Can Form Mini i-Motif Structures at Physiological pH.
J. Am. Chem. Soc., 139, 2017
|
|
5NYT
 
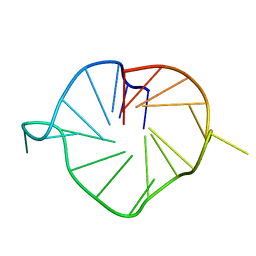 | | M2 G-quadruplex 20 wt% ethylene glycol | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
5NYU
 
 | | M2 G-quadruplex 10 wt% PEG8000 | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
1ONT
 
 | | NMDA RECEPTOR ANTAGONIST, CONANTOKIN-T, NMR, 17 STRUCTURES | | Descriptor: | CONANTOKIN-T | | Authors: | Skjaerbaek, N, Nielsen, K.J, Lewis, R.J, Alewood, P.F, Craik, D.J. | | Deposit date: | 1996-08-27 | | Release date: | 1997-09-04 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Determination of the solution structures of conantokin-G and conantokin-T by CD and NMR spectroscopy.
J.Biol.Chem., 272, 1997
|
|
4R3D
 
 | | Crystal structure of MERS Coronavirus papain like protease | | Descriptor: | Non-structural protein 3, ZINC ION | | Authors: | Kong, L.Y, Wang, Q, Ming, Z.H, Shaw, N, Sun, Y.N, Yan, L.M, Lou, Z.Y, Rao, Z.H. | | Deposit date: | 2014-08-15 | | Release date: | 2015-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.818 Å) | | Cite: | The Crystal Structures of Middle East Respiratory Syndrome Coronavirus Papain-like Protease and Its Complex with an Inhibitor
To be Published
|
|
5NYS
 
 | | M2 G-quadruplex dilute solution | | Descriptor: | DNA (5'-D(*TP*AP*GP*GP*GP*AP*CP*GP*GP*GP*CP*GP*GP*GP*CP*AP*GP*GP*GP*T)-3') | | Authors: | Trajkovski, M, Plavec, J, Endoh, T, Tateishi-Karimata, H, Sugimoto, N. | | Deposit date: | 2017-05-11 | | Release date: | 2018-04-11 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | Pursuing origins of (poly)ethylene glycol-induced G-quadruplex structural modulations.
Nucleic Acids Res., 46, 2018
|
|
4YKF
 
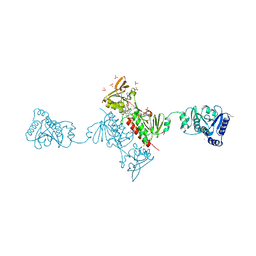 | | Crystal Structure of the Alkylhydroperoxide Reductase subunit F (AhpF) with NADH from Escherichia coli | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Alkyl hydroperoxide reductase subunit F, CADMIUM ION, ... | | Authors: | Kamariah, N, Manimekalai, M.S.S, Gruber, G, Eisenhaber, F, Eisenhaber, B. | | Deposit date: | 2015-03-04 | | Release date: | 2015-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystallographic and solution studies of NAD(+)- and NADH-bound alkylhydroperoxide reductase subunit F (AhpF) from Escherichia coli provide insight into sequential enzymatic steps
Biochim.Biophys.Acta, 1847, 2015
|
|
4R76
 
 | |
4R9V
 
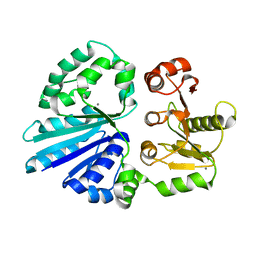 | | Crystal structure of sialyltransferase from photobacterium damselae, residues 113-497 corresponding to the gt-b domain | | Descriptor: | CALCIUM ION, Sialyltransferase 0160 | | Authors: | Li, Y, Huynh, N, Chen, X, Fisher, A.J. | | Deposit date: | 2014-09-08 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of sialyltransferase from Photobacterium damselae.
Febs Lett., 588, 2014
|
|
4RC2
 
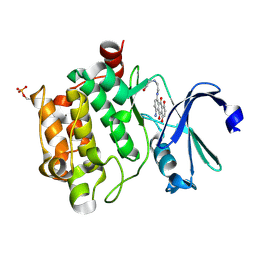 | |
5MB7
 
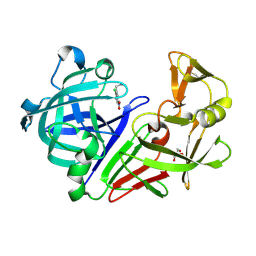 | |
1NEP
 
 | | Crystal Structure Analysis of the Bovine NPC2 (Niemann-Pick C2) Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Epididymal secretory protein E1, PHOSPHATE ION | | Authors: | Friedland, N, Liou, H.-L, Lobel, P, Stock, A.M. | | Deposit date: | 2002-12-11 | | Release date: | 2003-01-28 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a Cholesterol-binding Protein Deficient in Niemann-Pick Type C2 Disease
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
4RF2
 
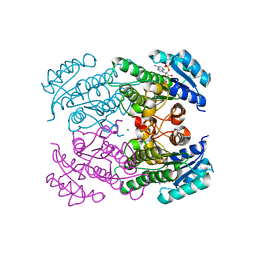 | |
5OON
 
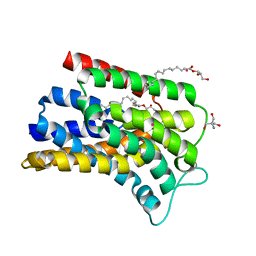 | | Structure of Undecaprenyl-Pyrophosphate Phosphatase, BacA | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MERCURY (II) ION, ... | | Authors: | Huang, C.-Y, Olieric, V, Warshamanage, R, Wang, M, Howe, N, Ghachi, M.E.I, Weichert, D, Kerff, F, Stansfeld, P, Touze, T, Caffrey, M. | | Deposit date: | 2017-08-08 | | Release date: | 2018-03-21 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of undecaprenyl-pyrophosphate phosphatase and its role in peptidoglycan biosynthesis.
Nat Commun, 9, 2018
|
|
4R5E
 
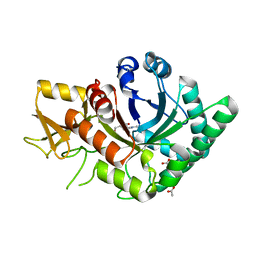 | | Crystal Structure of Family GH18 Chitinase from Cycas revoluta a Complex with Allosamidin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-allopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-allopyranose, ACETATE ION, ALLOSAMIZOLINE, ... | | Authors: | Umemoto, N, Numata, T, Ohnuma, T, Fukamizo, T. | | Deposit date: | 2014-08-21 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures and inhibitor binding properties of plant class V chitinases: the cycad enzyme exhibits unique structural and functional features.
Plant J., 82, 2015
|
|
1N71
 
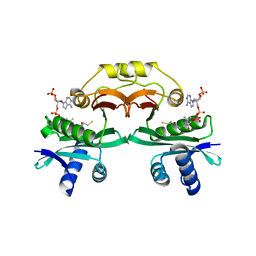 | | Crystal structure of aminoglycoside 6'-acetyltransferase type Ii in complex with coenzyme A | | Descriptor: | COENZYME A, SULFATE ION, aac(6')-Ii | | Authors: | Burk, D.L, Ghuman, N, Wybenga-Groot, L.E, Berghuis, A.M. | | Deposit date: | 2002-11-12 | | Release date: | 2003-03-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray structure of the AAC(6')-Ii antibiotic resistance enzyme at 1.8 A resolution; examination of oligomeric arrangements in GNAT superfamily members
Protein Sci., 12, 2003
|
|
4R5Z
 
 | | Crystal structure of Rv3772 encoded aminotransferase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Nasir, N, Anant, A, Vyas, R, Biswal, B.K. | | Deposit date: | 2014-08-22 | | Release date: | 2015-08-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structures of Mycobacterium tuberculosis HspAT and ArAT reveal structural basis of their distinct substrate specificities
Sci Rep, 6, 2016
|
|
4R6H
 
 | | Crystal structure of putative binding protein msme from bacillus subtilis subsp. subtilis str. 168, target efi-510764, an open conformation | | Descriptor: | CHLORIDE ION, Solute binding protein MsmE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al obaidi, N, Chamala, S, Attonito, J.D, Scott glenn, A, Chowdhury, S, Lafleur, J, Siedel, R.D, Hillerich, B, Love, J, Whalen, K.L, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-08-25 | | Release date: | 2014-09-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of Transporter Msme from Bacillus Subtilis, Target Efi-510764
To be Published
|
|
1NEE
 
 | | Structure of archaeal translation factor aIF2beta from Methanobacterium thermoautrophicum | | Descriptor: | Probable translation initiation factor 2 beta subunit, ZINC ION | | Authors: | Gutierrez, P, Trempe, J.F, Siddiqui, N, Arrowsmith, C, Gehring, K. | | Deposit date: | 2002-12-11 | | Release date: | 2004-03-09 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the archaeal translation initiation factor aIF2beta from Methanobacterium thermoautotrophicum: Implications for translation initiation.
Protein Sci., 13, 2004
|
|
5OQL
 
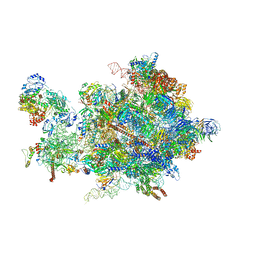 | | Cryo-EM structure of the 90S pre-ribosome from Chaetomium thermophilum | | Descriptor: | 35S rRNA, 40S ribosomal protein S1, 40S ribosomal protein S11-like protein, ... | | Authors: | Cheng, J, Kellner, N, Berninghausen, O, Hurt, E, Beckmann, R. | | Deposit date: | 2017-08-12 | | Release date: | 2017-10-11 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | 3.2- angstrom -resolution structure of the 90S preribosome before A1 pre-rRNA cleavage.
Nat. Struct. Mol. Biol., 24, 2017
|
|
4R7U
 
 | | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin | | Descriptor: | SODIUM ION, TETRAETHYLENE GLYCOL, UDP-N-acetylglucosamine 1-carboxyvinyltransferase, ... | | Authors: | Nocek, B, Maltseva, N, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-28 | | Release date: | 2014-09-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase from Vibrio cholerae in complex with substrate UDP-N-acetylglucosamine and the drug fosfomycin
To be Published
|
|
