4DC5
 
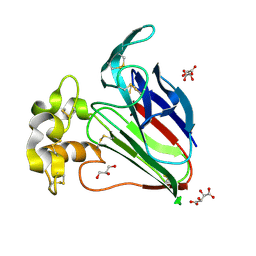 | | Crystal Structure of Thaumatin Unexposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Mulichak, A.M, Becker, M, Kissick, D.J, Keefe, L.J, Fischetti, R.F, Simpson, G.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2Q3F
 
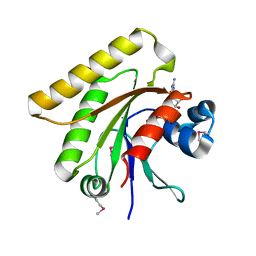 | | X-ray crystal structure of putative human Ras-related GTP binding D in complex with GMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related GTP-binding protein D | | Authors: | Mulichak, A.M, Rabeh, W.M, Tempel, W, Nedyalkova, L, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Keefe, L.J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-30 | | Release date: | 2007-06-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of human Ras-related GTP-binding D.
To be published
|
|
1N7H
 
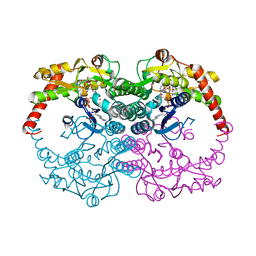 | | Crystal Structure of GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of the MUR1 GDP-mannose
4,6-dehydratase from A. thaliana:
Implications for ligand binding and specificity.
Biochemistry, 41, 2002
|
|
1N7G
 
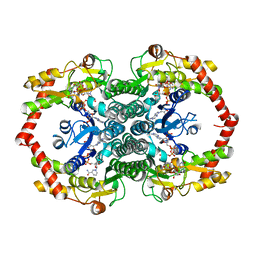 | | Crystal Structure of the GDP-mannose 4,6-dehydratase ternary complex with NADPH and GDP-rhamnose. | | Descriptor: | GDP-D-mannose-4,6-dehydratase, GUANOSINE-5'-DIPHOSPHATE-RHAMNOSE, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Mulichak, A.M, Bonin, C.P, Reiter, W.-D, Garavito, R.M. | | Deposit date: | 2002-11-14 | | Release date: | 2003-01-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the MUR1 GDP-mannose 4,6-dehydratase
from A. thaliana: Implications for ligand binding and
specificity.
Biochemistry, 41, 2002
|
|
1BG3
 
 | |
1BDG
 
 | |
4DC6
 
 | | Crystal Structure of Thaumatin Exposed to Excessive SONICC Imaging Laser Dose. | | Descriptor: | GLYCEROL, L(+)-TARTARIC ACID, Thaumatin I | | Authors: | Mulichak, A.M, Becker, M, Kissick, D.J, Keefe, L.J, Fischetti, R.F, Simpson, G.J. | | Deposit date: | 2012-01-17 | | Release date: | 2013-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Towards protein-crystal centering using second-harmonic generation (SHG) microscopy.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4HL7
 
 | | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae | | Descriptor: | Nicotinate phosphoribosyltransferase, SULFATE ION | | Authors: | Mulichak, A.M, Sauder, J.M, Keefe, L.J, Burley, S.K, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2012-10-16 | | Release date: | 2012-11-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of nicotinate phosphoribosyltransferase (target NYSGR-026035) from Vibrio cholerae
To be Published
|
|
1IIR
 
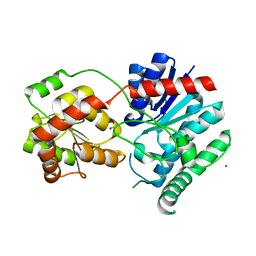 | | Crystal Structure of UDP-glucosyltransferase GtfB | | Descriptor: | MAGNESIUM ION, SULFATE ION, glycosyltransferase GtfB | | Authors: | Mulichak, A.M, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2001-04-24 | | Release date: | 2001-07-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the UDP-glucosyltransferase GtfB that modifies the heptapeptide aglycone in the biosynthesis of vancomycin group antibiotics.
Structure, 9, 2001
|
|
1IVQ
 
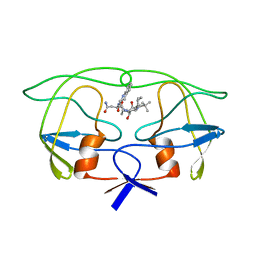 | |
1IVP
 
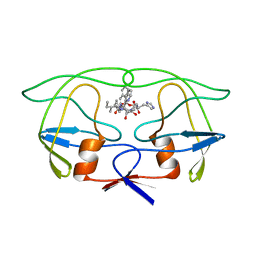 | | THE CRYSTALLOGRAPHIC STRUCTURE OF THE PROTEASE FROM HUMAN IMMUNODEFICIENCY VIRUS TYPE 2 WITH TWO SYNTHETIC PEPTIDIC TRANSITION STATE ANALOG INHIBITORS | | Descriptor: | 4-[(2R)-3-{[(1S,2S,3R,4S)-1-(cyclohexylmethyl)-2,3-dihydroxy-5-methyl-4-({(1S,2R)-2-methyl-1-[(pyridin-2-ylmethyl)carba moyl]butyl}carbamoyl)hexyl]amino}-2-{[(naphthalen-1-yloxy)acetyl]amino}-3-oxopropyl]-1H-imidazol-3-ium, HIV-2 PROTEASE | | Authors: | Mulichak, A.M, Watenpaugh, K.D. | | Deposit date: | 1993-03-18 | | Release date: | 1993-07-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystallographic structure of the protease from human immunodeficiency virus type 2 with two synthetic peptidic transition state analog inhibitors.
J.Biol.Chem., 268, 1993
|
|
2HPE
 
 | |
2HPF
 
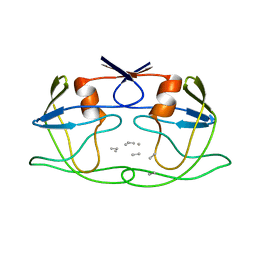 | |
1PN3
 
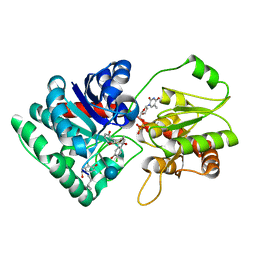 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and the acceptor substrate DVV. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-12 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1QRR
 
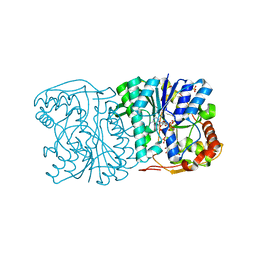 | | CRYSTAL STRUCTURE OF SQD1 PROTEIN COMPLEX WITH NAD AND UDP-GLUCOSE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, URIDINE-5'-DIPHOSPHATE-GLUCOSE, ... | | Authors: | Mulichak, A.M, Theisen, M.J, Essigmann, B, Benning, C, Garavito, R.M. | | Deposit date: | 1999-06-15 | | Release date: | 1999-11-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of SQD1, an enzyme involved in the biosynthesis of the plant sulfolipid headgroup donor UDP-sulfoquinovose.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
1RRV
 
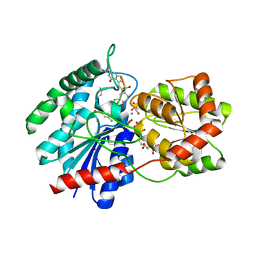 | | X-ray crystal structure of TDP-vancosaminyltransferase GtfD as a complex with TDP and the natural substrate, desvancosaminyl vancomycin. | | Descriptor: | DESVANCOSAMINYL VANCOMYCIN, GLYCEROL, GLYCOSYLTRANSFERASE GTFD, ... | | Authors: | Mulichak, A.M, Lu, W, Losey, H.C, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-12-09 | | Release date: | 2004-05-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Vancosaminyltransferase Gtfd from the Vancomycin Biosynthetic Pathway: Interactions with Acceptor and Nucleotide Ligands
Biochemistry, 43, 2004
|
|
1PNV
 
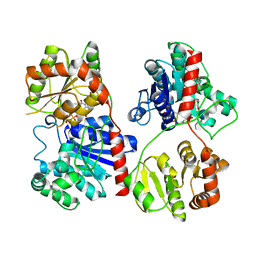 | | Crystal Structure of TDP-epi-Vancosaminyltransferase GtfA in complexes with TDP and Vancomycin | | Descriptor: | GLYCOSYLTRANSFERASE GTFA, THYMIDINE-5'-DIPHOSPHATE, VANCOMYCIN, ... | | Authors: | Mulichak, A.M, Losey, H.C, Lu, W, Wawrzak, Z, Walsh, C.T, Garavito, R.M. | | Deposit date: | 2003-06-13 | | Release date: | 2003-08-12 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the Tdp-Epi-Vancosaminyltransferase Gtfa from the Chloroeremomycin Biosynthetic Pathway.
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1TA9
 
 | |
2GQF
 
 | | Crystal structure of flavoprotein HI0933 from Haemophilus influenzae Rd | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Hypothetical protein HI0933, SULFATE ION | | Authors: | Mulichak, A.M, Patskovsky, Y, Keefe, L.J, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2006-04-20 | | Release date: | 2006-05-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of hypothetical flavoprotein HI0933 from Haemophilus influenzae Rd.
To be Published
|
|
1Z6G
 
 | | Crystal structure of guanylate kinase from Plasmodium falciparum | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, SULFATE ION, guanylate kinase | | Authors: | Mulichak, A.M, Lew, J, Artz, J, Choe, J, Walker, J.R, Zhao, Y, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Hui, R, Gao, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2005-03-22 | | Release date: | 2005-04-05 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Genome-scale protein expression and structural biology of Plasmodium falciparum and related Apicomplexan organisms.
Mol.Biochem.Parasitol., 151, 2007
|
|
4UPJ
 
 | |
6UPJ
 
 | | HIV-2 PROTEASE/U99294 COMPLEX | | Descriptor: | 6,7,8,9-TETRAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOHEPTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
1UPJ
 
 | |
5UPJ
 
 | | HIV-2 PROTEASE/U99283 COMPLEX | | Descriptor: | 5,6,7,8,9,10-HEXAHYDRO-4-HYDROXY-3-(1-PHENYLPROPYL)CYCLOOCTA[B]PYRAN-2-ONE, HIV-2 PROTEASE | | Authors: | Watenpaugh, K.D, Mulichak, A.M, Finzel, B.C. | | Deposit date: | 1996-12-10 | | Release date: | 1997-04-21 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Use of medium-sized cycloalkyl rings to enhance secondary binding: discovery of a new class of human immunodeficiency virus (HIV) protease inhibitors.
J.Med.Chem., 38, 1995
|
|
3UPJ
 
 | |
