1K1B
 
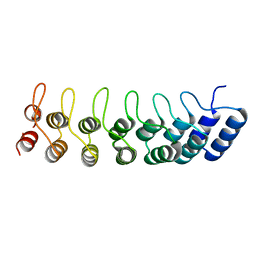 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
1K1A
 
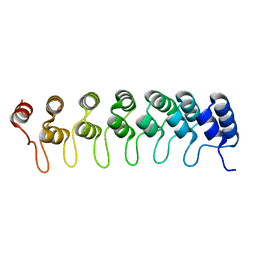 | | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family | | Descriptor: | B-cell lymphoma 3-encoded protein | | Authors: | Michel, F, Soler-Lopez, M, Petosa, C, Cramer, P, Siebenlist, U, Mueller, C.W. | | Deposit date: | 2001-09-24 | | Release date: | 2001-11-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Crystal structure of the ankyrin repeat domain of Bcl-3: a unique member of the IkappaB protein family.
EMBO J., 20, 2001
|
|
7OB9
 
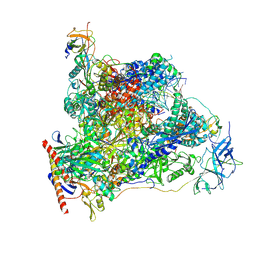 | | Cryo-EM structure of human RNA Polymerase I in elongation state | | Descriptor: | DNA non-template strand, DNA template strand, DNA-directed RNA polymerase I subunit RPA1, ... | | Authors: | Misiaszek, A.D, Girbig, M, Mueller, C.W. | | Deposit date: | 2021-04-21 | | Release date: | 2021-12-08 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryo-EM structures of human RNA polymerase I.
Nat.Struct.Mol.Biol., 28, 2021
|
|
7QBA
 
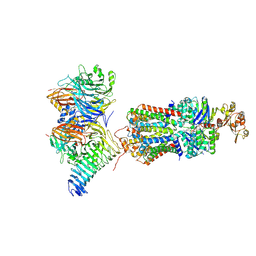 | | CryoEM structure of the ABC transporter NosDFY complexed with nitrous oxide reductase NosZ | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Zipfel, S, Mueller, C, Topitsch, A, Lutz, M, Zhang, L, Einsle, O. | | Deposit date: | 2021-11-18 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Molecular interplay of an assembly machinery for nitrous oxide reductase.
Nature, 608, 2022
|
|
6RKZ
 
 | | Recombinant Pseudomonas stutzeri nitrous oxide reductase, form II | | Descriptor: | (MU-4-SULFIDO)-TETRA-NUCLEAR COPPER ION, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, ... | | Authors: | Zhang, L, Wuest, A, Prasser, B, Mueller, C, Einsle, O. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Functional assembly of nitrous oxide reductase provides insights into copper site maturation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
1BVO
 
 | | DORSAL HOMOLOGUE GAMBIF1 BOUND TO DNA | | Descriptor: | DNA DUPLEX, TRANSCRIPTION FACTOR GAMBIF1 | | Authors: | Cramer, P, Varrot, A, Barillas-Mury, C, Kafatos, F.C, Mueller, C.W. | | Deposit date: | 1998-09-16 | | Release date: | 1999-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of the specificity domain of the Dorsal homologue Gambif1 bound to DNA.
Structure Fold.Des., 7, 1999
|
|
4D4Q
 
 | | Crystal Structure of Kti13/AtS1 | | Descriptor: | PROTEIN ATS1 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.395 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
5FLV
 
 | | Crystal structure of NKX2-5 and TBX5 bound to the Nppa promoter region | | Descriptor: | 5'-D(*AP*CP*CP*AP*CP*TP*TP*CP*AP*AP*AP*GP*GP*TP *GP*TP*GP*AP*GP*AP*AP*G)-3', 5'-D(*TP*CP*TP*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP *TP*GP*AP*AP*GP*TP*GP*G)-3', HOMEOBOX PROTEIN NKX-2.5, ... | | Authors: | Stirnimann, C.U, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-10-28 | | Release date: | 2016-02-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis.
Cell(Cambridge,Mass.), 164, 2016
|
|
6HBD
 
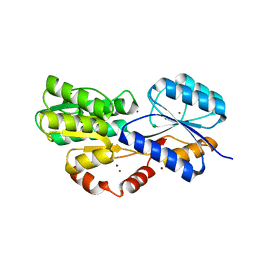 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Galactofuranose | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION, beta-D-galactofuranose | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HBM
 
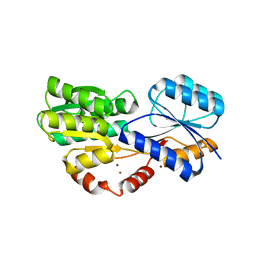 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with alpha-L-arabinofuranose | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION, alpha-L-arabinofuranose | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-10 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HN5
 
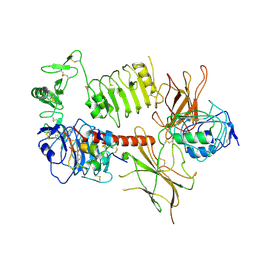 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "upper" membrane-distal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin, ... | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
6HN4
 
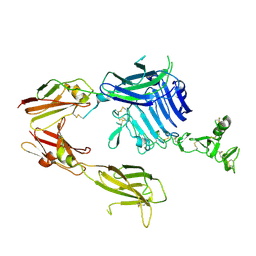 | | Leucine-zippered human insulin receptor ectodomain with single bound insulin - "lower" membrane-proximal part | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor,Insulin receptor,General control protein GCN4 | | Authors: | Weis, F, Menting, J.G, Margetts, M.B, Chan, S.J, Xu, Y, Tennagels, N, Wohlfart, P, Langer, T, Mueller, C.W, Dreyer, M.K, Lawrence, M.C. | | Deposit date: | 2018-09-14 | | Release date: | 2018-11-21 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | The signalling conformation of the insulin receptor ectodomain.
Nat Commun, 9, 2018
|
|
6HB0
 
 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis | | Descriptor: | ABC transporter periplasmic-binding protein YtfQ, ZINC ION | | Authors: | Li, M, Mueller, C, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-08-09 | | Release date: | 2019-05-01 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
2XYI
 
 | | Crystal Structure of Nurf55 in complex with a H4 peptide | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTONE H4, PROBABLE HISTONE-BINDING PROTEIN CAF1, ... | | Authors: | Stirnimann, C.U, Nowak, A.J, Mueller, C.W. | | Deposit date: | 2010-11-17 | | Release date: | 2011-05-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Chromatin-Modifying Complex Component Nurf55/P55 Associates with Histones H3, H4 and Polycomb Repressive Complex 2 Subunit Su(Z)12 Through Partially Overlapping Binding Sites.
J.Biol.Chem., 286, 2011
|
|
6HYH
 
 | | Crystal structure of MSMEG_1712 from Mycobacterium smegmatis in complex with Beta-D-Fucofuranose | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, ZINC ION, beta-D-fucofuranose | | Authors: | Li, M, Mueller, C, Zhang, L, Einsle, O, Jessen-Trefzer, C. | | Deposit date: | 2018-10-21 | | Release date: | 2019-05-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detection and Characterization of a Mycobacterial L-Arabinofuranose ABC Transporter Identified with a Rapid Lipoproteomics Protocol.
Cell Chem Biol, 26, 2019
|
|
6HGC
 
 | | Structure of Calypso in complex with DEUBAD of ASX | | Descriptor: | Polycomb protein Asx, Ubiquitin carboxyl-terminal hydrolase calypso,Ubiquitin carboxyl-terminal hydrolase calypso | | Authors: | De, I, Chittock, E.C, Groetsch, H, Miller, T.C.R, McCarthy, A.A, Mueller, C.W. | | Deposit date: | 2018-08-23 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Structural Basis for the Activation of the Deubiquitinase Calypso by the Polycomb Protein ASX.
Structure, 27, 2019
|
|
5AIM
 
 | | Crystal structure of T138 central eWH domain | | Descriptor: | GLYCEROL, TRANSCRIPTION FACTOR TAU 138 KDA SUBUNIT | | Authors: | Male, G, Glatt, S, Mueller, C.W. | | Deposit date: | 2015-02-16 | | Release date: | 2015-06-24 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.401 Å) | | Cite: | Architecture of TFIIIC and its role in RNA polymerase III pre-initiation complex assembly.
Nat Commun, 6, 2015
|
|
4D4O
 
 | | Crystal Structure of the Kti11 Kti13 heterodimer Spacegroup P64 | | Descriptor: | FE (III) ION, PROTEIN ATS1, DIPHTHAMIDE BIOSYNTHESIS PROTEIN 3, ... | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
4D4P
 
 | | Crystal Structure of the Kti11 Kti13 heterodimer Spacegroup P65 | | Descriptor: | FE (III) ION, PROTEIN ATS1, DIPHTHAMIDE BIOSYNTHESIS PROTEIN 3, ... | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2014-10-30 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.999 Å) | | Cite: | Structure of the Kti11/Kti13 Heterodimer and its Double Role in Modifications of tRNA and Eukaryotic Elongation Factor 2.
Structure, 23, 2015
|
|
5A48
 
 | |
5A4A
 
 | | Crystal structure of the OSK domain of Drosophila Oskar | | Descriptor: | MATERNAL EFFECT PROTEIN OSKAR, SULFATE ION | | Authors: | Jeske, M, Glatt, S, Ephrussi, A, Mueller, C.W. | | Deposit date: | 2015-06-05 | | Release date: | 2015-07-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.699 Å) | | Cite: | The Crystal Structure of the Drosophila Germline Inducer Oskar Identifies Two Domains with Distinct Vasa Helicase-and RNA-Binding Activities.
Cell Rep., 12, 2015
|
|
5AIO
 
 | |
6RL0
 
 | | Recombinant Pseudomonas stutzeri nitrous oxide reductase, form I | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Zhang, L, Wuest, A, Prasser, B, Mueller, C, Einsle, O. | | Deposit date: | 2019-04-30 | | Release date: | 2019-06-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Functional assembly of nitrous oxide reductase provides insights into copper site maturation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2X6V
 
 | | Crystal structure of human TBX5 in the DNA-bound and DNA-free form | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 5'-D(*TP*AP*AP*GP*GP*TP*GP*TP*GP*AP*GP)-3', 5'-D(*TP*CP*TP*CP*AP*CP*AP*CP*CP*TP*TP)-3', ... | | Authors: | Ptchelkine, D, Stirnimann, C.U, Grimm, C, Mueller, C.W. | | Deposit date: | 2010-02-22 | | Release date: | 2010-04-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Basis of Tbx5-DNA Recognition: The T-Box Domain in its DNA-Bound and -Unbound Form.
J.Mol.Biol., 400, 2010
|
|
4A8J
 
 | | Crystal Structure of the Elongator subcomplex Elp456 | | Descriptor: | Elongator complex protein 4, Elongator complex protein 5, Elongator complex protein 6 | | Authors: | Glatt, S, Mueller, C.W. | | Deposit date: | 2011-11-21 | | Release date: | 2012-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Elongator Subcomplex Elp456 is a Hexameric Reca-Like ATPase.
Nat.Struct.Mol.Biol., 19, 2012
|
|
